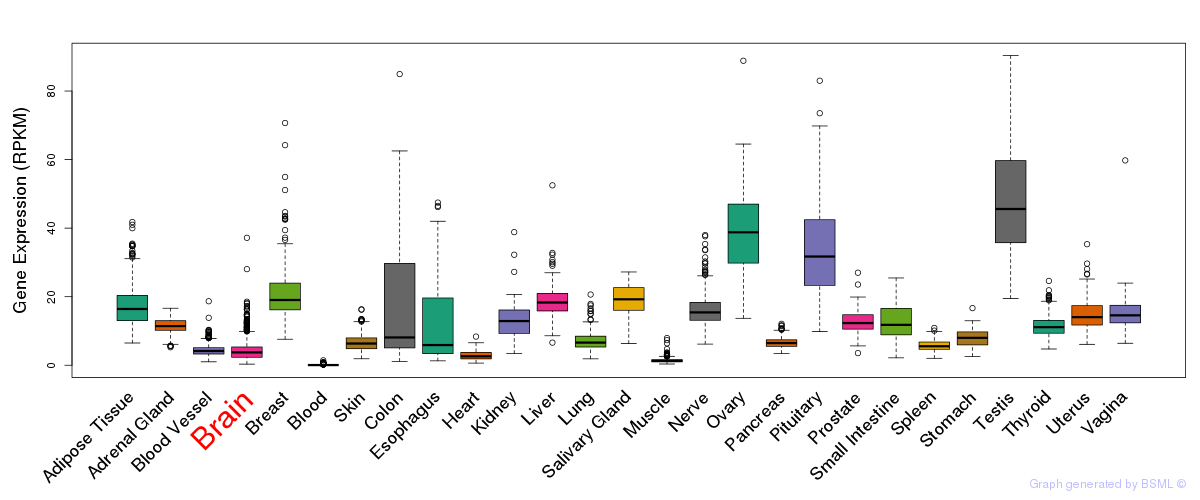
| ONKEN UVEAL MELANOMA DN
| 526 | 357 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN
| 349 | 157 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN
| 384 | 230 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL QUIESCENT DN
| 47 | 32 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT DN
| 95 | 57 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP
| 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN
| 805 | 505 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL DN
| 101 | 66 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN
| 1024 | 594 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS MODERATELY UP
| 109 | 69 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP
| 344 | 180 | All SZGR 2.0 genes in this pathway |
| BALDWIN PRKCI TARGETS UP
| 35 | 26 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS
| 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS
| 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS
| 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS
| 882 | 572 | All SZGR 2.0 genes in this pathway |
| BECKER TAMOXIFEN RESISTANCE DN
| 52 | 37 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL
| 254 | 164 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP
| 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN
| 504 | 323 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 8
| 86 | 57 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN
| 91 | 64 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS DN
| 135 | 88 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN
| 128 | 93 | All SZGR 2.0 genes in this pathway |
| TSENG ADIPOGENIC POTENTIAL DN
| 46 | 28 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4
| 307 | 185 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN
| 216 | 143 | All SZGR 2.0 genes in this pathway |
| WANG NEOPLASTIC TRANSFORMATION BY CCND1 MYC
| 21 | 14 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN
| 435 | 289 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2
| 356 | 214 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK
| 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN
| 668 | 419 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN
| 193 | 112 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT UP
| 166 | 105 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE
| 721 | 492 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN
| 321 | 200 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN
| 784 | 464 | All SZGR 2.0 genes in this pathway |
| BOSCO EPITHELIAL DIFFERENTIATION MODULE
| 69 | 31 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP
| 297 | 194 | All SZGR 2.0 genes in this pathway |
 eQTL annotation
eQTL annotation