| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN
| 634 | 384 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP
| 783 | 507 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER UP
| 206 | 111 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP
| 485 | 293 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN
| 406 | 230 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN
| 180 | 101 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL UP
| 95 | 64 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP
| 418 | 263 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN
| 240 | 171 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP
| 544 | 308 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP
| 431 | 237 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS DN
| 136 | 94 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS UP
| 126 | 72 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP
| 191 | 128 | All SZGR 2.0 genes in this pathway |
| MISSIAGLIA REGULATED BY METHYLATION UP
| 126 | 78 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP
| 271 | 175 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS DN
| 62 | 44 | All SZGR 2.0 genes in this pathway |
| OLSSON E2F3 TARGETS UP
| 28 | 14 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP
| 207 | 128 | All SZGR 2.0 genes in this pathway |
| DAWSON METHYLATED IN LYMPHOMA TCL1
| 59 | 45 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1
| 229 | 137 | All SZGR 2.0 genes in this pathway |
| ROPERO HDAC2 TARGETS
| 114 | 71 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP
| 77 | 60 | All SZGR 2.0 genes in this pathway |
| WU CELL MIGRATION
| 184 | 114 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS
| 734 | 436 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 60 HELA
| 46 | 32 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN
| 357 | 212 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP
| 223 | 140 | All SZGR 2.0 genes in this pathway |
| KENNY CTNNB1 TARGETS UP
| 50 | 30 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH CBFB MYH11 FUSION
| 52 | 32 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP
| 398 | 262 | All SZGR 2.0 genes in this pathway |
| SANA TNF SIGNALING UP
| 83 | 56 | All SZGR 2.0 genes in this pathway |
| OSAWA TNF TARGETS
| 10 | 6 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN
| 85 | 67 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP
| 183 | 111 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS
| 191 | 131 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP
| 101 | 76 | All SZGR 2.0 genes in this pathway |
| URS ADIPOCYTE DIFFERENTIATION UP
| 74 | 51 | All SZGR 2.0 genes in this pathway |
| ZHOU TNF SIGNALING 4HR
| 54 | 36 | All SZGR 2.0 genes in this pathway |
| LIAN LIPA TARGETS 6M
| 74 | 47 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS DN
| 64 | 44 | All SZGR 2.0 genes in this pathway |
| LIAN LIPA TARGETS 3M
| 59 | 36 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP
| 487 | 303 | All SZGR 2.0 genes in this pathway |
| GEISS RESPONSE TO DSRNA UP
| 38 | 29 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN
| 287 | 208 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS UP
| 113 | 71 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A UP
| 81 | 52 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE
| 206 | 117 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN
| 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP
| 783 | 442 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP
| 393 | 244 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3
| 720 | 440 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP
| 282 | 183 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS DN
| 145 | 93 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS DN
| 97 | 53 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP
| 259 | 185 | All SZGR 2.0 genes in this pathway |
| HUPER BREAST BASAL VS LUMINAL DN
| 59 | 44 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP
| 165 | 118 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS UP
| 77 | 56 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION B
| 53 | 36 | All SZGR 2.0 genes in this pathway |
| PARK TRETINOIN RESPONSE AND PML RARA FUSION
| 30 | 21 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN
| 209 | 137 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE UP
| 91 | 63 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS FIBROBLAST UP
| 84 | 60 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK
| 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED
| 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3
| 269 | 159 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION
| 368 | 247 | All SZGR 2.0 genes in this pathway |
| TIAN TNF SIGNALING VIA NFKB
| 28 | 21 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION
| 456 | 287 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP
| 857 | 456 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP
| 397 | 206 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP
| 430 | 288 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN
| 882 | 538 | All SZGR 2.0 genes in this pathway |
| BILANGES RAPAMYCIN SENSITIVE GENES
| 39 | 20 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP
| 163 | 111 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D
| 882 | 506 | All SZGR 2.0 genes in this pathway |
| PHONG TNF TARGETS UP
| 63 | 43 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38
| 337 | 236 | All SZGR 2.0 genes in this pathway |
| ALTEMEIER RESPONSE TO LPS WITH MECHANICAL VENTILATION
| 128 | 81 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN
| 428 | 246 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL PROGENITOR UP
| 58 | 30 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY
| 1839 | 928 | All SZGR 2.0 genes in this pathway |
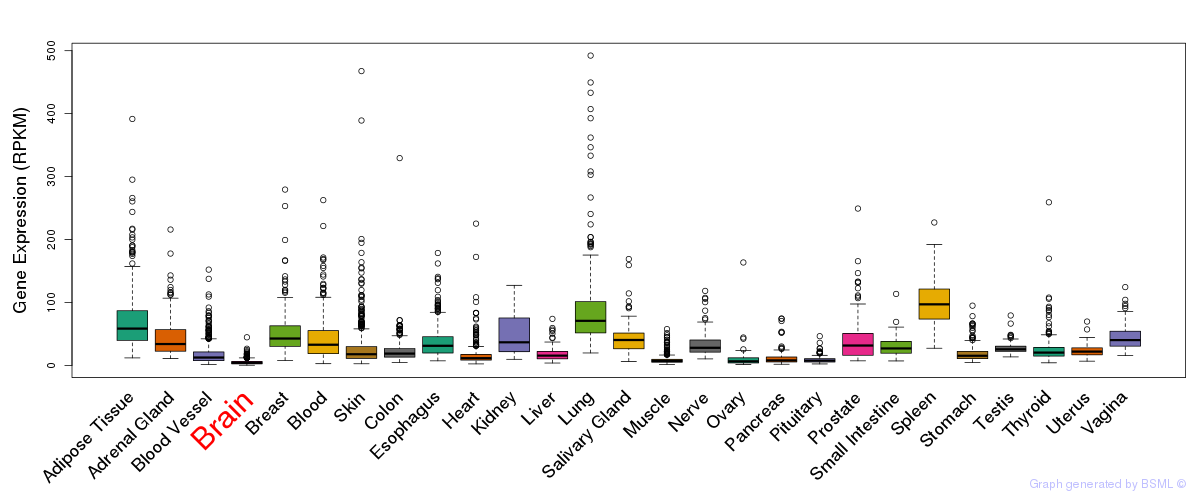
 eQTL annotation
eQTL annotation