Gene Page: TRPC6
Summary ?
| GeneID | 7225 |
| Symbol | TRPC6 |
| Synonyms | FSGS2|TRP6 |
| Description | transient receptor potential cation channel subfamily C member 6 |
| Reference | MIM:603652|HGNC:HGNC:12338|Ensembl:ENSG00000137672|HPRD:04710|Vega:OTTHUMG00000167483 |
| Gene type | protein-coding |
| Map location | 11q22.1 |
| Pascal p-value | 0.619 |
| Fetal beta | -0.421 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
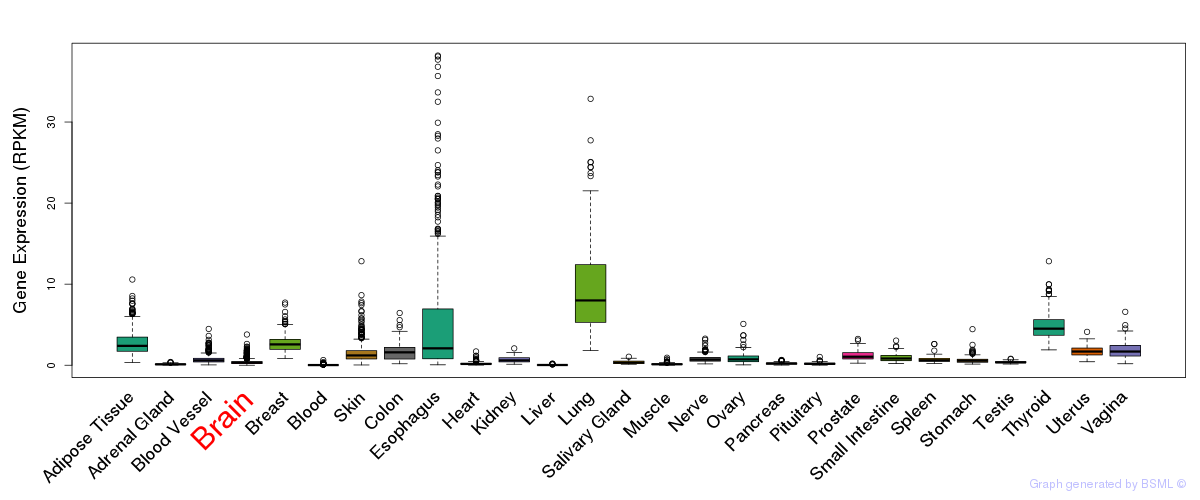
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CSTF1 | 0.90 | 0.90 |
| RNF34 | 0.90 | 0.89 |
| PRPF4 | 0.89 | 0.90 |
| SMU1 | 0.89 | 0.89 |
| WBP11 | 0.89 | 0.87 |
| KIAA0406 | 0.89 | 0.91 |
| SLBP | 0.89 | 0.86 |
| SLC30A5 | 0.88 | 0.87 |
| EFTUD2 | 0.88 | 0.89 |
| GORASP2 | 0.88 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.84 | -0.84 |
| AF347015.31 | -0.81 | -0.82 |
| AF347015.21 | -0.81 | -0.85 |
| AF347015.8 | -0.81 | -0.83 |
| MT-CYB | -0.80 | -0.81 |
| AF347015.33 | -0.80 | -0.81 |
| AF347015.27 | -0.79 | -0.81 |
| AF347015.2 | -0.77 | -0.79 |
| AF347015.15 | -0.76 | -0.81 |
| FXYD1 | -0.75 | -0.79 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 14761972 |
| ITPR1 | INSP3R1 | IP3R | IP3R1 | SCA15 | SCA16 | inositol 1,4,5-triphosphate receptor, type 1 | Affinity Capture-Western | BioGRID | 14505576 |
| ITPR3 | FLJ36205 | IP3R | IP3R3 | inositol 1,4,5-triphosphate receptor, type 3 | - | HPRD,BioGRID | 11290752 |
| MX1 | IFI-78K | IFI78 | MX | MxA | myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) | MxA interacts with TRPC6. | BIND | 15757897 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 14761972 |
| TRPC1 | HTRP-1 | MGC133334 | MGC133335 | TRP1 | transient receptor potential cation channel, subfamily C, member 1 | Affinity Capture-Western | BioGRID | 12857742 |
| TRPC2 | - | transient receptor potential cation channel, subfamily C, member 2 (pseudogene) | Affinity Capture-Western | BioGRID | 14699131 |
| TRPC3 | TRP3 | transient receptor potential cation channel, subfamily C, member 3 | - | HPRD,BioGRID | 12032305 |
| TRPC3 | TRP3 | transient receptor potential cation channel, subfamily C, member 3 | - | HPRD | 15623527 |
| TRPC4 | HTRP4 | MGC119570 | MGC119571 | MGC119572 | MGC119573 | TRP4 | transient receptor potential cation channel, subfamily C, member 4 | Affinity Capture-Western | BioGRID | 12857742 |
| TRPC5 | TRP5 | transient receptor potential cation channel, subfamily C, member 5 | Affinity Capture-Western | BioGRID | 12857742 |
| TRPC6 | FLJ11098 | FLJ14863 | FSGS2 | TRP6 | transient receptor potential cation channel, subfamily C, member 6 | - | HPRD,BioGRID | 12032305 |
| TRPC7 | TRP7 | transient receptor potential cation channel, subfamily C, member 7 | FRET | BioGRID | 12032305 |
| TRPM2 | EREG1 | KNP3 | LTRPC2 | MGC133383 | NUDT9H | NUDT9L1 | TRPC7 | transient receptor potential cation channel, subfamily M, member 2 | - | HPRD | 12032305 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID ENDOTHELIN PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID NEPHRIN NEPH1 PATHWAY | 31 | 24 | All SZGR 2.0 genes in this pathway |
| PID EPO PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID THROMBIN PAR1 PATHWAY | 43 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | 10 | 6 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Q SIGNALLING EVENTS | 184 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME EFFECTS OF PIP2 HYDROLYSIS | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME NETRIN1 SIGNALING | 41 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET HOMEOSTASIS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET CALCIUM HOMEOSTASIS | 18 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW UP | 162 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH DN | 180 | 110 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA DN | 181 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA PROGRESSION RISK | 74 | 44 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH VS LOW DN | 32 | 21 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1 | 229 | 137 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |