Gene Page: TUBB2A
Summary ?
| GeneID | 7280 |
| Symbol | TUBB2A |
| Synonyms | CDCBM5|TUBB|TUBB2 |
| Description | tubulin beta 2A class IIa |
| Reference | MIM:615101|HGNC:HGNC:12412|Ensembl:ENSG00000137267|HPRD:01852|Vega:OTTHUMG00000014135 |
| Gene type | protein-coding |
| Map location | 6p25 |
| Pascal p-value | 0.005 |
| Sherlock p-value | 0.886 |
| Fetal beta | 0.041 |
| Support | STRUCTURAL PLASTICITY G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0159 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
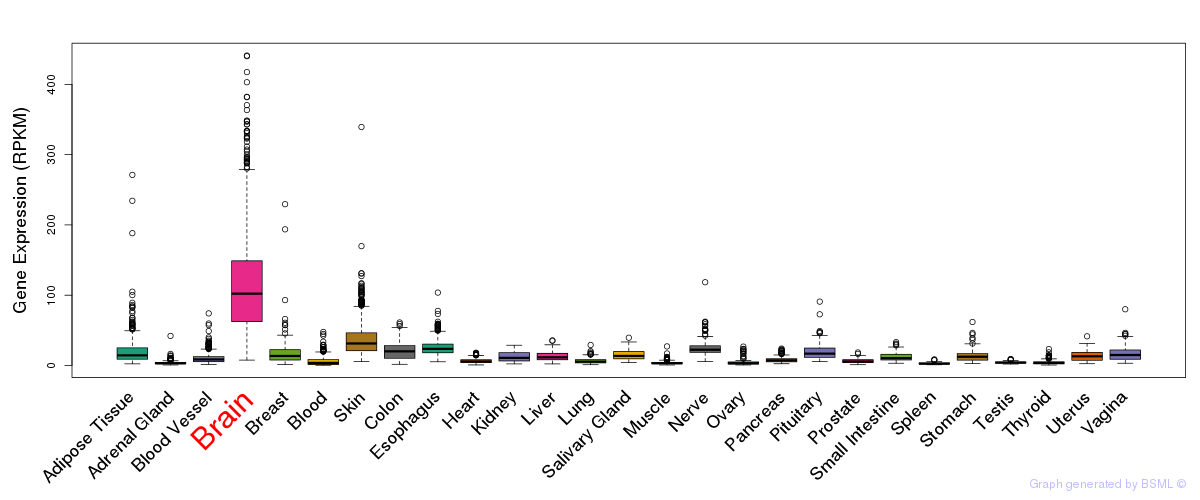
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| UBE2N | 0.93 | 0.91 |
| TMED2 | 0.91 | 0.91 |
| STRAP | 0.91 | 0.89 |
| ARMC1 | 0.91 | 0.90 |
| CNOT7 | 0.91 | 0.90 |
| PTGES3 | 0.90 | 0.88 |
| AZIN1 | 0.90 | 0.89 |
| TIPRL | 0.90 | 0.89 |
| ABCE1 | 0.90 | 0.89 |
| COPS3 | 0.90 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.71 | -0.70 |
| AF347015.2 | -0.68 | -0.69 |
| MT-CYB | -0.68 | -0.69 |
| AF347015.26 | -0.68 | -0.69 |
| AF347015.8 | -0.67 | -0.69 |
| AF347015.33 | -0.67 | -0.68 |
| AF347015.31 | -0.67 | -0.67 |
| FXYD1 | -0.65 | -0.71 |
| HIGD1B | -0.65 | -0.68 |
| GPT | -0.64 | -0.69 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003924 | GTPase activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 17373842 | |
| GO:0005525 | GTP binding | IEA | - | |
| GO:0005198 | structural molecule activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007018 | microtubule-based movement | IEA | - | |
| GO:0051258 | protein polymerization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005874 | microtubule | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0043234 | protein complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ALDOA | ALDA | MGC10942 | MGC17716 | MGC17767 | aldolase A, fructose-bisphosphate | - | HPRD | 8103323 |
| ARL15 | ARFRP2 | FLJ20051 | ADP-ribosylation factor-like 15 | Two-hybrid | BioGRID | 16169070 |
| ARL8B | ARL10C | FLJ10702 | Gie1 | ADP-ribosylation factor-like 8B | - | HPRD | 15331635 |
| C20orf20 | Eaf7 | FLJ10914 | MRG15BP | MRGBP | URCC4 | chromosome 20 open reading frame 20 | - | HPRD | 12963728 |
| CHGB | SCG1 | chromogranin B (secretogranin 1) | Two-hybrid | BioGRID | 16169070 |
| CRIPT | HSPC139 | cysteine-rich PDZ-binding protein | - | HPRD | 9581762 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | - | HPRD | 12884273 |
| GRM1 | GPRC1A | GRM1A | MGLUR1 | MGLUR1A | mGlu1 | glutamate receptor, metabotropic 1 | - | HPRD | 9886087 |
| LAMA4 | DKFZp686D23145 | LAMA3 | LAMA4*-1 | laminin, alpha 4 | Two-hybrid | BioGRID | 16169070 |
| MAP1A | FLJ77111 | MAP1L | MTAP1A | microtubule-associated protein 1A | - | HPRD | 11278895 |
| MAP3K10 | MLK2 | MST | mitogen-activated protein kinase kinase kinase 10 | - | HPRD | 9629920 |
| MAP3K7 | TAK1 | TGF1a | mitogen-activated protein kinase kinase kinase 7 | - | HPRD | 14743216 |
| MAP4 | DKFZp779A1753 | MGC8617 | microtubule-associated protein 4 | - | HPRD | 12855424 |
| MAP6 | FLJ41346 | KIAA1878 | MTAP6 | N-STOP | STOP | microtubule-associated protein 6 | - | HPRD | 8700896 |
| MAPT | DDPAC | FLJ31424 | FTDP-17 | MAPTL | MGC138549 | MSTD | MTBT1 | MTBT2 | PPND | TAU | microtubule-associated protein tau | - | HPRD | 9922135 |
| MARK4 | FLJ90097 | KIAA1860 | MARKL1 | Nbla00650 | MAP/microtubule affinity-regulating kinase 4 | - | HPRD | 14594945 |
| MX1 | IFI-78K | IFI78 | MX | MxA | myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) | - | HPRD | 1629950 |
| NCALD | MGC33870 | MGC74858 | neurocalcin delta | - | HPRD | 11964161 |
| NPHP1 | FLJ97602 | JBTS4 | NPH1 | SLSN1 | nephronophthisis 1 (juvenile) | - | HPRD | 12872123 |
| PAFAH1B3 | - | platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa | Two-hybrid | BioGRID | 16169070 |
| PIGT | CGI-06 | FLJ41596 | MGC8909 | NDAP | phosphatidylinositol glycan anchor biosynthesis, class T | - | HPRD | 12052837 |
| PSMD11 | MGC3844 | Rpn6 | S9 | p44.5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | Two-hybrid | BioGRID | 16169070 |
| PTPRS | PTPSIGMA | protein tyrosine phosphatase, receptor type, S | Two-hybrid | BioGRID | 16169070 |
| RAB8B | FLJ38125 | RAB8B, member RAS oncogene family | - | HPRD | 12639940 |
| RABAC1 | PRA1 | PRAF1 | YIP3 | Rab acceptor 1 (prenylated) | Two-hybrid | BioGRID | 16169070 |
| RASSF1 | 123F2 | NORE2A | RASSF1A | RDA32 | REH3P21 | Ras association (RalGDS/AF-6) domain family member 1 | - | HPRD | 14603253 |
| RIC8A | MGC104517 | MGC131931 | MGC148073 | MGC148074 | RIC8 | synembryn | resistance to inhibitors of cholinesterase 8 homolog A (C. elegans) | Two-hybrid | BioGRID | 16169070 |
| RPA2 | REPA2 | RPA32 | replication protein A2, 32kDa | Two-hybrid | BioGRID | 16169070 |
| RPLP1 | FLJ27448 | MGC5215 | P1 | RPP1 | ribosomal protein, large, P1 | Two-hybrid | BioGRID | 16169070 |
| SAT1 | DC21 | KFSD | SAT | SSAT | SSAT-1 | spermidine/spermine N1-acetyltransferase 1 | Two-hybrid | BioGRID | 16169070 |
| SDC3 | N-syndecan | SDCN | SYND3 | syndecan 3 | - | HPRD,BioGRID | 9553134 |
| SERPINB9 | CAP-3 | CAP3 | PI9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 | Two-hybrid | BioGRID | 16169070 |
| STXBP1 | EIEE4 | MUNC18-1 | UNC18 | hUNC18 | rbSec1 | syntaxin binding protein 1 | Affinity Capture-Western | BioGRID | 12963086 |
| SYT9 | FLJ45896 | synaptotagmin IX | - | HPRD | 12966166 |
| TAOK2 | KIAA0881 | MAP3K17 | PSK | PSK1 | TAO1 | TAO2 | TAO kinase 2 | - | HPRD | 12639963 |
| TBCD | KIAA0988 | tubulin folding cofactor D | - | HPRD | 10722852 |
| TGM2 | G-ALPHA-h | GNAH | TG2 | TGC | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | - | HPRD | 9973324 |
| TK1 | TK2 | thymidine kinase 1, soluble | Two-hybrid | BioGRID | 16169070 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD | 14743216 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GAP JUNCTION | 90 | 68 | All SZGR 2.0 genes in this pathway |
| KEGG PATHOGENIC ESCHERICHIA COLI INFECTION | 59 | 36 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSII PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSIII PATHWAY | 25 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | 28 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME PROTEIN FOLDING | 53 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | 22 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | 19 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA DN | 136 | 86 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE DN | 92 | 51 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA UP | 183 | 119 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER BASAL VS MESENCHYMAL DN | 50 | 36 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D UP | 157 | 91 | All SZGR 2.0 genes in this pathway |
| HUMMEL BURKITTS LYMPHOMA UP | 43 | 27 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL QUIESCENT UP | 87 | 45 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL DIVIDING UP | 57 | 33 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS DN | 136 | 94 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS DN | 142 | 95 | All SZGR 2.0 genes in this pathway |
| CASTELLANO NRAS TARGETS UP | 68 | 41 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2 | 127 | 92 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE LATE UP | 57 | 41 | All SZGR 2.0 genes in this pathway |
| WIEMANN TELOMERE SHORTENING AND CHRONIC LIVER DAMAGE UP | 8 | 5 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM2 | 153 | 102 | All SZGR 2.0 genes in this pathway |
| COWLING MYCN TARGETS | 43 | 27 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK UP | 63 | 48 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| RADMACHER AML PROGNOSIS | 78 | 52 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIC DAMAGE 24HR | 35 | 22 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| MCCLUNG DELTA FOSB TARGETS 2WK | 48 | 36 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL UP | 120 | 89 | All SZGR 2.0 genes in this pathway |
| SARTIPY NORMAL AT INSULIN RESISTANCE UP | 34 | 27 | All SZGR 2.0 genes in this pathway |
| ZHANG ANTIVIRAL RESPONSE TO RIBAVIRIN DN | 51 | 32 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR UP | 93 | 65 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G1 | 67 | 41 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 8HR | 39 | 31 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G2 | 30 | 18 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS | 47 | 28 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL UP | 45 | 26 | All SZGR 2.0 genes in this pathway |
| PODAR RESPONSE TO ADAPHOSTIN UP | 147 | 98 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION C | 69 | 49 | All SZGR 2.0 genes in this pathway |
| RAY TARGETS OF P210 BCR ABL FUSION UP | 18 | 13 | All SZGR 2.0 genes in this pathway |
| HOFMANN MYELODYSPLASTIC SYNDROM LOW RISK DN | 30 | 20 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS SIGNALING UP | 41 | 24 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 11 | 103 | 68 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G24 | 28 | 19 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN | 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 | 182 | 102 | All SZGR 2.0 genes in this pathway |
| RAGHAVACHARI PLATELET SPECIFIC GENES | 70 | 46 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 3 DN | 39 | 21 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE GENES | 90 | 54 | All SZGR 2.0 genes in this pathway |
| BILANGES RAPAMYCIN SENSITIVE VIA TSC1 AND TSC2 | 73 | 37 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 PARTIAL | 160 | 106 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL UP | 146 | 75 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-200bc/429 | 147 | 153 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-29 | 76 | 82 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.