Gene Page: XK
Summary ?
| GeneID | 7504 |
| Symbol | XK |
| Synonyms | KX|MCLDS|NA|NAC|X1k|XKR1 |
| Description | X-linked Kx blood group |
| Reference | MIM:314850|HGNC:HGNC:12811|Ensembl:ENSG00000047597|HPRD:02444|Vega:OTTHUMG00000033171 |
| Gene type | protein-coding |
| Map location | Xp21.1 |
| Sherlock p-value | 0.031 |
| Fetal beta | -3.225 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
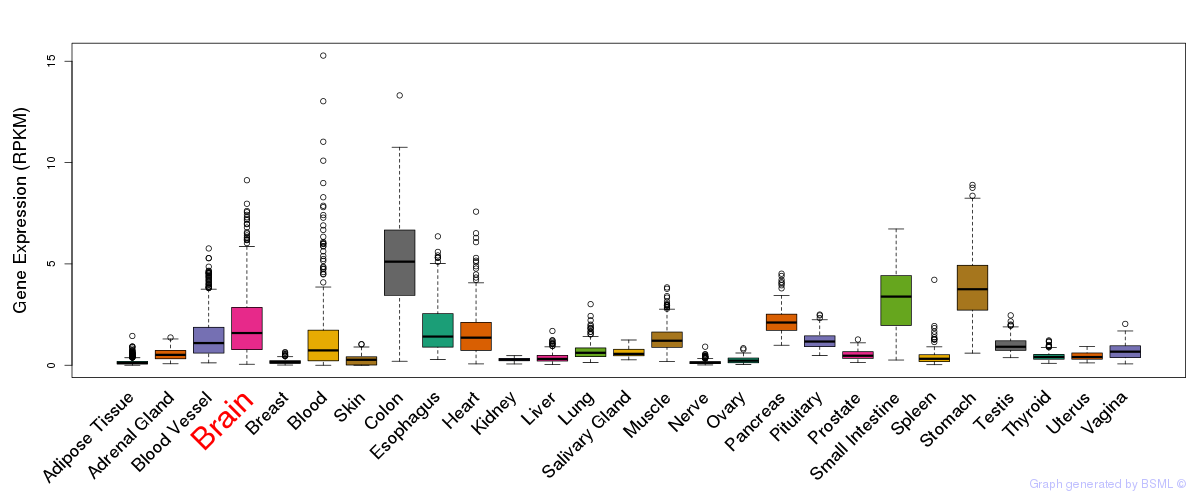
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SCML4 | 0.82 | 0.35 |
| ZIC1 | 0.82 | 0.69 |
| ID4 | 0.81 | 0.50 |
| ZIC2 | 0.77 | 0.59 |
| FZD10 | 0.76 | 0.47 |
| GBX2 | 0.75 | 0.32 |
| ZIC4 | 0.75 | 0.49 |
| EPHA1 | 0.74 | 0.14 |
| RCAN3 | 0.73 | 0.41 |
| PROX1 | 0.73 | 0.36 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.25 | -0.34 |
| CA4 | -0.23 | -0.22 |
| TMEM54 | -0.23 | -0.22 |
| HLA-F | -0.23 | -0.22 |
| SLC26A4 | -0.23 | -0.25 |
| AF347015.27 | -0.23 | -0.29 |
| AF347015.33 | -0.22 | -0.31 |
| COX7A1 | -0.22 | -0.43 |
| EPHX4 | -0.22 | -0.24 |
| CA11 | -0.22 | -0.28 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 7737196 | |
| GO:0005215 | transporter activity | TAS | 8004674 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006810 | transport | TAS | 8004674 | |
| GO:0006865 | amino acid transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | TAS | 9647734 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL QUIESCENT UP | 87 | 45 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT UP | 181 | 101 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING DN | 87 | 49 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| OKUMURA INFLAMMATORY RESPONSE LPS | 183 | 115 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| SCHURINGA STAT5A TARGETS UP | 21 | 13 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART UP | 103 | 69 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C3 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| KRASNOSELSKAYA ILF3 TARGETS DN | 46 | 38 | All SZGR 2.0 genes in this pathway |
| TUOMISTO TUMOR SUPPRESSION BY COL13A1 UP | 18 | 9 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL DN | 127 | 75 | All SZGR 2.0 genes in this pathway |
| ZHAN LATE DIFFERENTIATION GENES UP | 33 | 24 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G23 UP | 52 | 35 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 DN | 170 | 105 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER WITH EPCAM UP | 53 | 25 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL UP | 193 | 95 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS UP | 139 | 93 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS UP | 266 | 142 | All SZGR 2.0 genes in this pathway |