Gene Page: SF1
Summary ?
| GeneID | 7536 |
| Symbol | SF1 |
| Synonyms | BBP|D11S636|MBBP|ZCCHC25|ZFM1|ZNF162 |
| Description | splicing factor 1 |
| Reference | MIM:601516|HGNC:HGNC:12950|Ensembl:ENSG00000168066|HPRD:03306|Vega:OTTHUMG00000066833 |
| Gene type | protein-coding |
| Map location | 11q13 |
| Pascal p-value | 0.422 |
| Fetal beta | 0.893 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04619046 | 11 | 64541730 | SF1 | 2.476E-4 | -0.604 | 0.037 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6576747 | chr1 | 85530213 | SF1 | 7536 | 0.19 | trans | ||
| rs17025528 | chr1 | 216043152 | SF1 | 7536 | 0.1 | trans | ||
| rs12997903 | chr2 | 1952083 | SF1 | 7536 | 0.08 | trans | ||
| rs6733654 | chr2 | 1963270 | SF1 | 7536 | 0.04 | trans | ||
| rs891877 | chr2 | 2043348 | SF1 | 7536 | 0.03 | trans | ||
| rs1395163 | chr3 | 31028951 | SF1 | 7536 | 0 | trans | ||
| rs17029291 | chr3 | 32402138 | SF1 | 7536 | 1.254E-4 | trans | ||
| rs6446109 | chr3 | 60076988 | SF1 | 7536 | 0.07 | trans | ||
| rs624496 | chr3 | 120506736 | SF1 | 7536 | 0.02 | trans | ||
| rs16870339 | chr5 | 2705169 | SF1 | 7536 | 0.01 | trans | ||
| rs4617108 | chr7 | 49423831 | SF1 | 7536 | 0.13 | trans | ||
| rs17160578 | chr7 | 85672108 | SF1 | 7536 | 0.13 | trans | ||
| rs722970 | chr7 | 85803264 | SF1 | 7536 | 0.07 | trans | ||
| rs17324099 | chr7 | 85856288 | SF1 | 7536 | 0.13 | trans | ||
| snp_a-1904613 | 0 | SF1 | 7536 | 0.02 | trans | |||
| rs1831517 | chr9 | 2513405 | SF1 | 7536 | 0.07 | trans | ||
| rs7864672 | chr9 | 31068069 | SF1 | 7536 | 0.03 | trans | ||
| rs11259374 | chr10 | 14917473 | SF1 | 7536 | 0.08 | trans | ||
| rs4607971 | chr10 | 19407890 | SF1 | 7536 | 0.04 | trans | ||
| rs12569493 | chr10 | 74886042 | SF1 | 7536 | 0.07 | trans | ||
| rs16930362 | chr10 | 74890320 | SF1 | 7536 | 0.11 | trans | ||
| rs909288 | chr10 | 124257417 | SF1 | 7536 | 0.12 | trans | ||
| rs11021829 | chr11 | 11416216 | SF1 | 7536 | 0.15 | trans | ||
| rs17133022 | chr11 | 98980829 | SF1 | 7536 | 0.17 | trans | ||
| rs9554597 | chr13 | 100117238 | SF1 | 7536 | 0.17 | trans | ||
| rs10145685 | chr14 | 22794892 | SF1 | 7536 | 0.02 | trans | ||
| rs17793827 | chr14 | 22797560 | SF1 | 7536 | 0.01 | trans | ||
| rs10136383 | chr14 | 22801078 | SF1 | 7536 | 0.01 | trans | ||
| rs2455956 | chr14 | 43524686 | SF1 | 7536 | 0.04 | trans | ||
| rs2254834 | chr14 | 43596184 | SF1 | 7536 | 0.01 | trans | ||
| rs1954396 | chr14 | 43601835 | SF1 | 7536 | 1.31E-4 | trans | ||
| rs9925128 | chr16 | 79102822 | SF1 | 7536 | 0.11 | trans | ||
| rs1761450 | chr19 | 54818034 | SF1 | 7536 | 0.03 | trans | ||
| rs16996418 | chr20 | 9778936 | SF1 | 7536 | 0.04 | trans | ||
| rs16996420 | chr20 | 9780867 | SF1 | 7536 | 0.01 | trans | ||
| rs12157904 | chr22 | 37982011 | SF1 | 7536 | 0.03 | trans |
Section II. Transcriptome annotation
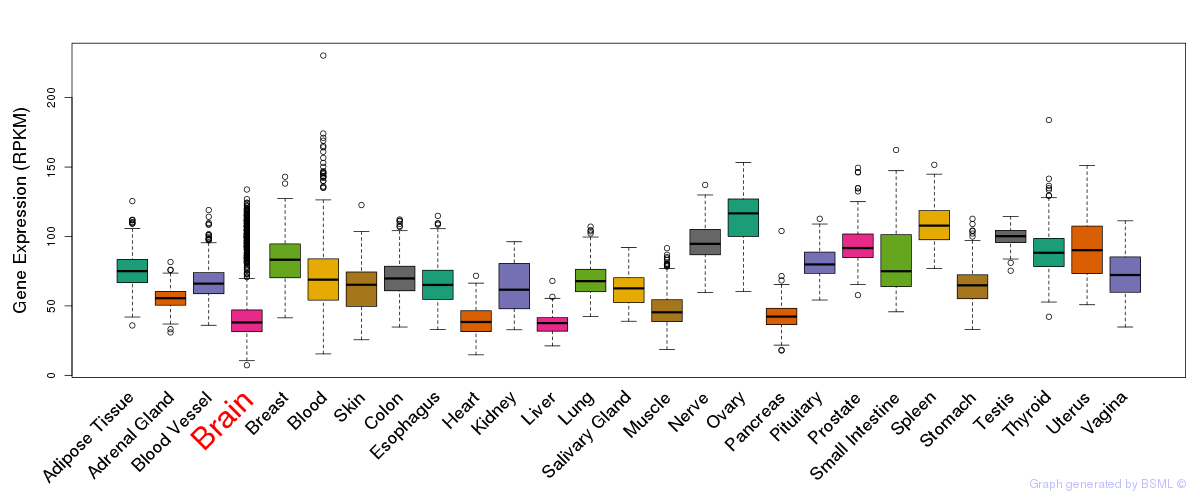
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CHD4 | 0.92 | 0.92 |
| CRKRS | 0.91 | 0.93 |
| ZNF264 | 0.91 | 0.93 |
| LIG3 | 0.91 | 0.90 |
| ZNF740 | 0.91 | 0.90 |
| KDM3B | 0.91 | 0.92 |
| CYTSA | 0.90 | 0.92 |
| GCN1L1 | 0.90 | 0.90 |
| KPNB1 | 0.90 | 0.92 |
| ZXDC | 0.90 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.72 | -0.83 |
| FXYD1 | -0.70 | -0.81 |
| IFI27 | -0.70 | -0.81 |
| MT-CO2 | -0.69 | -0.81 |
| AF347015.27 | -0.68 | -0.79 |
| HIGD1B | -0.68 | -0.81 |
| AF347015.33 | -0.67 | -0.77 |
| AF347015.21 | -0.67 | -0.85 |
| S100B | -0.66 | -0.75 |
| CXCL14 | -0.66 | -0.79 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | - | HPRD,BioGRID | 8668206 |
| DDX17 | DKFZp761H2016 | P72 | RH70 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | Two-hybrid | BioGRID | 16189514 |
| EWSR1 | EWS | Ewing sarcoma breakpoint region 1 | - | HPRD,BioGRID | 9660765 |
| FUS | CHOP | FUS-CHOP | FUS1 | TLS | TLS/CHOP | hnRNP-P2 | fusion (involved in t(12;16) in malignant liposarcoma) | - | HPRD,BioGRID | 9660765 |
| PLSCR1 | MMTRA1B | phospholipid scramblase 1 | Two-hybrid | BioGRID | 16189514 |
| PRPF40A | FBP-11 | FBP11 | FLAF1 | FLJ20585 | FNBP3 | HIP10 | HYPA | NY-REN-6 | PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) | - | HPRD | 15456888 |
| RBPMS | HERMES | RNA binding protein with multiple splicing | Two-hybrid | BioGRID | 16189514 |
| TAF15 | Npl3 | RBP56 | TAF2N | TAFII68 | hTAFII68 | TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa | - | HPRD,BioGRID | 9660765 |
| TCERG1 | CA150 | MGC133200 | TAF2S | transcription elongation regulator 1 | - | HPRD,BioGRID | 11604498 |
| TRIM23 | ARD1 | ARFD1 | RNF46 | tripartite motif-containing 23 | Two-hybrid | BioGRID | 16189514 |
| U2AF2 | U2AF65 | U2 small nuclear RNA auxiliary factor 2 | - | HPRD,BioGRID | 9150140 |9512519 |
| U2AF2 | U2AF65 | U2 small nuclear RNA auxiliary factor 2 | U2AF65 interacts with an unspecified isoform of SF1 (mBBP). | BIND | 9512519 |
| WWP2 | AIP2 | WWp2-like | WW domain containing E3 ubiquitin protein ligase 2 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA UP | 177 | 110 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA DN | 104 | 59 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| WOOD EBV EBNA1 TARGETS UP | 110 | 71 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 DN | 66 | 38 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER DN | 160 | 110 | All SZGR 2.0 genes in this pathway |
| BROCKE APOPTOSIS REVERSED BY IL6 | 144 | 98 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR UP | 151 | 100 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY TBH AND H2O2 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR UP | 111 | 68 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| MATZUK MALE REPRODUCTION SERTOLI | 28 | 23 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION | 130 | 87 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 36HR | 152 | 88 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |