Gene Page: CACNA1C
Summary ?
| GeneID | 775 |
| Symbol | CACNA1C |
| Synonyms | CACH2|CACN2|CACNL1A1|CCHL1A1|CaV1.2|LQT8|TS |
| Description | calcium voltage-gated channel subunit alpha1 C |
| Reference | MIM:114205|HGNC:HGNC:1390|Ensembl:ENSG00000151067|HPRD:00246|Vega:OTTHUMG00000150243 |
| Gene type | protein-coding |
| Map location | 12p13.3 |
| Pascal p-value | 1E-12 |
| Sherlock p-value | 0.003 |
| Fetal beta | -1.205 |
| DMG | 2 (# studies) |
| eGene | Cerebellum |
| Support | EXCITABILITY SEROTONIN G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGC128 | Genome-wide Association Study | A multi-stage schizophrenia GWAS of up to 36,989 cases and 113,075 controls. Reported by the Schizophrenia Working Group of PGC. 128 independent associations spanning 108 loci | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:Ripke_2013 | Genome-wide Association Study | Multi-stage GWAS, Sweden population and PGC2. 24 leading SNPs | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0047 |
Section I. Genetics and epigenetics annotation
 CV:PGC128
CV:PGC128
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs2007044 | chr12 | 2344960 | AG | 2.625E-17 | intronic | CACNA1C | |
| rs2239063 | chr12 | 2511831 | AC | 5.389E-9 | intronic | CACNA1C |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25519930 | 12 | 2161640 | CACNA1C | 5.628E-4 | 0.402 | 0.049 | DMG:Wockner_2014 |
| cg12282391 | 12 | 2162491 | CACNA1C | 9.08E-9 | -0.017 | 4.12E-6 | DMG:Jaffe_2016 |
| cg01398181 | 12 | 2279968 | CACNA1C | 7.8E-8 | 0.018 | 1.84E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
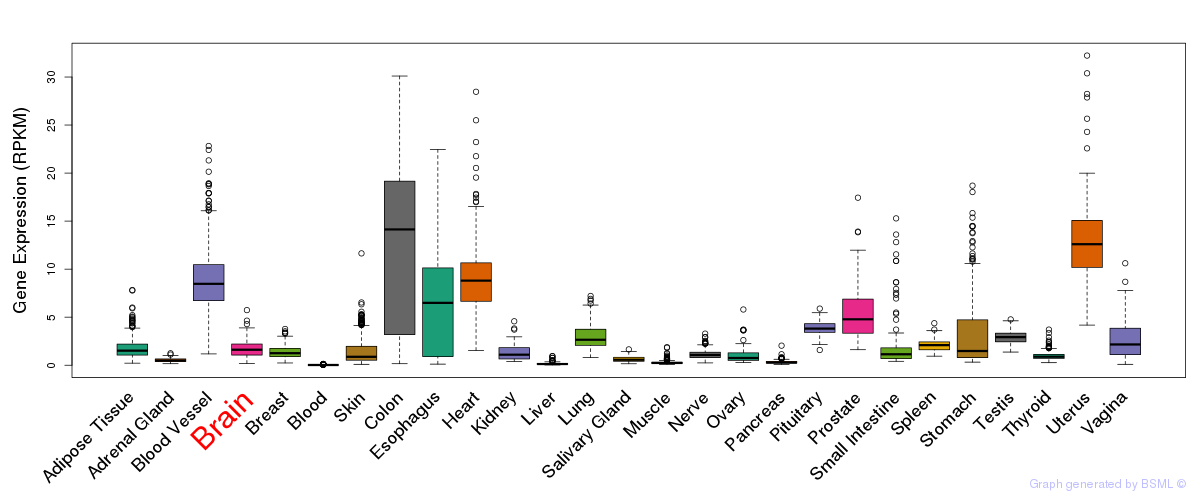
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CYP46A1 | 0.80 | 0.85 |
| TMEM38A | 0.75 | 0.83 |
| GRASP | 0.75 | 0.81 |
| ADAMTS8 | 0.74 | 0.76 |
| FBXL16 | 0.73 | 0.75 |
| STIM1 | 0.73 | 0.73 |
| STAT6 | 0.73 | 0.81 |
| DDN | 0.72 | 0.75 |
| PACSIN1 | 0.72 | 0.79 |
| CTSB | 0.71 | 0.67 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1949 | -0.58 | -0.55 |
| TUBB2B | -0.58 | -0.59 |
| TRAF4 | -0.56 | -0.64 |
| ZNF311 | -0.55 | -0.56 |
| DYNLT1 | -0.55 | -0.70 |
| PPAPDC1B | -0.55 | -0.52 |
| ZNF300 | -0.55 | -0.51 |
| RBMX2 | -0.55 | -0.69 |
| TIGD1 | -0.55 | -0.55 |
| YBX1 | -0.54 | -0.57 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005516 | calmodulin binding | IPI | 15140941 | |
| GO:0005244 | voltage-gated ion channel activity | IEA | - | |
| GO:0005245 | voltage-gated calcium channel activity | IEA | - | |
| GO:0005216 | ion channel activity | IEA | - | |
| GO:0015270 | dihydropyridine-sensitive calcium channel activity | IDA | 8392192 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007204 | elevation of cytosolic calcium ion concentration | IDA | 12130699 | |
| GO:0006816 | calcium ion transport | IEA | - | |
| GO:0006811 | ion transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0014069 | postsynaptic density | IDA | Synap (GO term level: 10) | 14140941 |
| GO:0005737 | cytoplasm | IDA | 11206130 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IDA | 8392192 | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IDA | 11206130 | |
| GO:0005891 | voltage-gated calcium channel complex | IDA | 12130699 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG CARDIAC MUSCLE CONTRACTION | 80 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY | 101 | 77 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE II DIABETES MELLITUS | 47 | 41 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS DN | 120 | 69 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 UP | 134 | 85 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION UP | 271 | 165 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 DN | 180 | 116 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 1695 | 1701 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-129-5p | 6702 | 6708 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-140 | 1330 | 1336 | m8 | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-153 | 1393 | 1399 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-155 | 1448 | 1454 | 1A | hsa-miR-155 | UUAAUGCUAAUCGUGAUAGGGG |
| miR-19 | 373 | 380 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-200bc/429 | 1594 | 1600 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-25/32/92/363/367 | 1406 | 1412 | 1A | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-26 | 1573 | 1580 | 1A,m8 | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-320 | 1737 | 1743 | 1A | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-33 | 1403 | 1410 | 1A,m8 | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-342 | 6621 | 6628 | 1A,m8 | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
| miR-377 | 6620 | 6626 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-381 | 1684 | 1691 | 1A,m8 | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-448 | 1392 | 1399 | 1A,m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-494 | 1549 | 1555 | m8 | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
| miR-496 | 1700 | 1707 | 1A,m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-505 | 1556 | 1562 | 1A | hsa-miR-505 | GUCAACACUUGCUGGUUUCCUC |
| miR-96 | 1634 | 1640 | m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.