Gene Page: CCDC28B
Summary ?
| GeneID | 79140 |
| Symbol | CCDC28B |
| Synonyms | - |
| Description | coiled-coil domain containing 28B |
| Reference | MIM:610162|HGNC:HGNC:28163|Ensembl:ENSG00000160050|HPRD:14422|Vega:OTTHUMG00000005738 |
| Gene type | protein-coding |
| Map location | 1p36.11-p34.2 |
| Pascal p-value | 0.177 |
| Sherlock p-value | 0.11 |
| Fetal beta | 1.182 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13299148 | 1 | 32170433 | CCDC28B | 0.004 | 3.812 | DMG:vanEijk_2014 | |
| cg10694959 | 1 | 32573640 | CCDC28B | 0.002 | 2.902 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
General gene expression (GTEx)
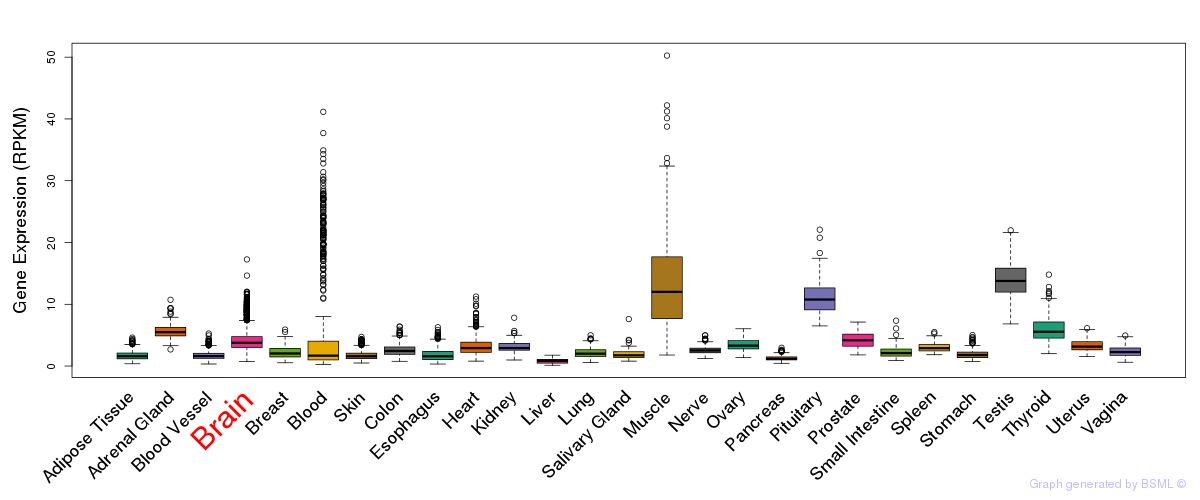
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZSCAN2 | 0.95 | 0.88 |
| BRD1 | 0.95 | 0.90 |
| KDM4A | 0.94 | 0.88 |
| PHF2 | 0.94 | 0.90 |
| PATZ1 | 0.94 | 0.89 |
| GPR161 | 0.94 | 0.84 |
| UBTD2 | 0.94 | 0.89 |
| CNOT6 | 0.94 | 0.88 |
| ZBTB5 | 0.94 | 0.91 |
| KDM5B | 0.94 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.72 | -0.76 |
| HLA-F | -0.70 | -0.73 |
| PTH1R | -0.69 | -0.73 |
| FBXO2 | -0.69 | -0.66 |
| AF347015.31 | -0.69 | -0.84 |
| AF347015.27 | -0.68 | -0.81 |
| AIFM3 | -0.68 | -0.70 |
| ALDOC | -0.68 | -0.67 |
| CA4 | -0.68 | -0.74 |
| TNFSF12 | -0.68 | -0.68 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| SCHLINGEMANN SKIN CARCINOGENESIS TPA DN | 29 | 15 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A DN | 159 | 105 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| KRIEG KDM3A TARGETS NOT HYPOXIA | 208 | 107 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |