Gene Page: CAMK2A
Summary ?
| GeneID | 815 |
| Symbol | CAMK2A |
| Synonyms | CAMKA |
| Description | calcium/calmodulin-dependent protein kinase II alpha |
| Reference | MIM:114078|HGNC:HGNC:1460|Ensembl:ENSG00000070808|HPRD:06532|Vega:OTTHUMG00000134281 |
| Gene type | protein-coding |
| Map location | 5q32 |
| Pascal p-value | 0.321 |
| Sherlock p-value | 0.533 |
| DMG | 1 (# studies) |
| eGene | Meta |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION SEROTONIN G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_PocklingtonH1 G2Cdb.human_Synaptosome G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01718 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00459 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1782 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07622079 | 5 | 149658453 | CAMK2A | 5.948E-4 | 0.452 | 0.05 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
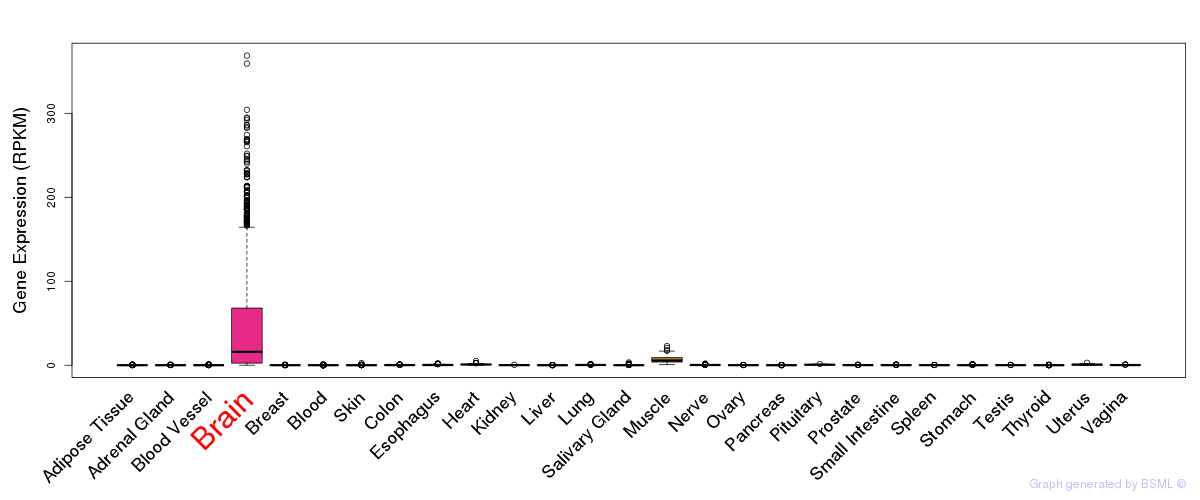
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CCT8 | 0.95 | 0.94 |
| ARMCX1 | 0.94 | 0.94 |
| ASNSD1 | 0.94 | 0.95 |
| ZUFSP | 0.94 | 0.94 |
| MRPL44 | 0.94 | 0.95 |
| TCP1 | 0.93 | 0.92 |
| TSG101 | 0.93 | 0.93 |
| CCT6A | 0.93 | 0.92 |
| IGBP1 | 0.93 | 0.93 |
| CCT4 | 0.93 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.77 | -0.86 |
| AF347015.33 | -0.77 | -0.86 |
| AF347015.27 | -0.76 | -0.86 |
| MT-CYB | -0.76 | -0.87 |
| AF347015.8 | -0.76 | -0.87 |
| AF347015.15 | -0.75 | -0.87 |
| AF347015.31 | -0.74 | -0.83 |
| AF347015.26 | -0.73 | -0.88 |
| HLA-F | -0.73 | -0.81 |
| AF347015.2 | -0.73 | -0.86 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005516 | calmodulin binding | IEA | - | |
| GO:0005524 | ATP binding | IC | 17052756 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004683 | calmodulin-dependent protein kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IDA | 17052756 | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0051092 | positive regulation of NF-kappaB transcription factor activity | IMP | 17052756 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005954 | calcium- and calmodulin-dependent protein kinase complex | TAS | 11264466 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTN4 | ACTININ-4 | DKFZp686K23158 | FSGS | FSGS1 | actinin, alpha 4 | CAMK2A (CaMKII-alpha) interacts with ACTN4 (alpha-Actinin 4). | BIND | 11160423 |
| ACTN4 | ACTININ-4 | DKFZp686K23158 | FSGS | FSGS1 | actinin, alpha 4 | Two-hybrid | BioGRID | 11160423 |
| ATF1 | EWS-ATF1 | FUS/ATF-1 | TREB36 | activating transcription factor 1 | Biochemical Activity | BioGRID | 8663317 |
| CALM3 | PHKD | PHKD3 | calmodulin 3 (phosphorylase kinase, delta) | Affinity Capture-MS | BioGRID | 17353931 |
| CAMK2B | CAM2 | CAMK2 | CAMKB | MGC29528 | calcium/calmodulin-dependent protein kinase II beta | Affinity Capture-MS | BioGRID | 17353931 |
| CAMK2D | CAMKD | DKFZp686G23119 | DKFZp686I2288 | MGC44911 | calcium/calmodulin-dependent protein kinase II delta | Affinity Capture-MS | BioGRID | 17353931 |
| CAMK2N2 | CAM-KIIN | CAMKIIN | calcium/calmodulin-dependent protein kinase II inhibitor 2 | - | HPRD | 9724800 |
| CDK5R1 | CDK5P35 | CDK5R | MGC33831 | NCK5A | p23 | p25 | p35 | p35nck5a | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | - | HPRD,BioGRID | 12223541 |
| CHMP5 | C9orf83 | CGI-34 | HSPC177 | PNAS-2 | SNF7DC2 | chromatin modifying protein 5 | Affinity Capture-MS | BioGRID | 17353931 |
| CREB1 | CREB | MGC9284 | cAMP responsive element binding protein 1 | Biochemical Activity | BioGRID | 8663317 |
| DAPK2 | DRP-1 | MGC119312 | death-associated protein kinase 2 | - | HPRD | 10629061 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | - | HPRD,BioGRID | 12933808 |
| DLG3 | KIAA1232 | MRX | MRX90 | NE-Dlg | NEDLG | SAP102 | discs, large homolog 3 (neuroendocrine-dlg, Drosophila) | Affinity Capture-MS | BioGRID | 17353931 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 10862698 |
| GRIN2B | MGC142178 | MGC142180 | NMDAR2B | NR2B | hNR3 | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | - | HPRD,BioGRID | 11459059 |
| ITPKA | IP3KA | inositol 1,4,5-trisphosphate 3-kinase A | - | HPRD,BioGRID | 11468283 |
| LRRC7 | DKFZp686I1147 | KIAA1365 | MGC144918 | leucine rich repeat containing 7 | - | HPRD | 11160423 |
| LRRC7 | DKFZp686I1147 | KIAA1365 | MGC144918 | leucine rich repeat containing 7 | LRRC7 (Densin-180) interacts with CAMK2A (CaMKII-alpha). This interaction was modeled on a demonstrated interaction between rat LRCC7 and human CAMK2A. | BIND | 11160423 |
| PARK2 | AR-JP | LPRS2 | PDJ | PRKN | Parkinson disease (autosomal recessive, juvenile) 2, parkin | Affinity Capture-Western | BioGRID | 11679592 |
| RIMS1 | CORD7 | KIAA0340 | MGC167823 | MGC176677 | RAB3IP2 | RIM | RIM1 | regulating synaptic membrane exocytosis 1 | Biochemical Activity | BioGRID | 12871946 |
| SYNGAP1 | DKFZp761G1421 | KIAA1938 | RASA1 | RASA5 | SYNGAP | synaptic Ras GTPase activating protein 1 homolog (rat) | - | HPRD | 11278737 |12951199 |
| TANC1 | KIAA1728 | ROLSB | TANC | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | - | HPRD | 15673434 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ERBB SIGNALING PATHWAY | 87 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG OLFACTORY TRANSDUCTION | 389 | 85 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY | 101 | 77 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOGENESIS | 102 | 80 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BIOPEPTIDES PATHWAY | 45 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CACAM PATHWAY | 16 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PGC1A PATHWAY | 26 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA STATHMIN PATHWAY | 19 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CREB PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| ST WNT CA2 CYCLIC GMP PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| PID WNT NONCANONICAL PATHWAY | 32 | 26 | All SZGR 2.0 genes in this pathway |
| PID NETRIN PATHWAY | 32 | 27 | All SZGR 2.0 genes in this pathway |
| PID IFNG PATHWAY | 40 | 34 | All SZGR 2.0 genes in this pathway |
| PID PI3K PLC TRK PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| PID FGF PATHWAY | 55 | 37 | All SZGR 2.0 genes in this pathway |
| PID TGFBR PATHWAY | 55 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAFFICKING OF AMPA RECEPTORS | 28 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | 17 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | 37 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | 33 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON GAMMA SIGNALING | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| MCCLUNG DELTA FOSB TARGETS 8WK | 47 | 38 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAINE REWARD 5D | 79 | 62 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAIN REWARD 4WK | 75 | 47 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| LEIN NEURON MARKERS | 69 | 45 | All SZGR 2.0 genes in this pathway |
| LEIN LOCALIZED TO DISTAL AND PROXIMAL DENDRITES | 17 | 12 | All SZGR 2.0 genes in this pathway |
| POS HISTAMINE RESPONSE NETWORK | 32 | 22 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES HCP WITH H3 UNMETHYLATED | 63 | 31 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3 UNMETHYLATED | 228 | 119 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-142-5p | 1499 | 1506 | 1A,m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-148/152 | 1479 | 1485 | m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-26 | 3100 | 3106 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-27 | 3094 | 3100 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-331 | 2732 | 2738 | 1A | hsa-miR-331brain | GCCCCUGGGCCUAUCCUAGAA |
| miR-338 | 88 | 94 | 1A | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| miR-495 | 3024 | 3030 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.