Gene Page: CAPZA1
Summary ?
| GeneID | 829 |
| Symbol | CAPZA1 |
| Synonyms | CAPPA1|CAPZ|CAZ1 |
| Description | capping actin protein of muscle Z-line alpha subunit 1 |
| Reference | MIM:601580|HGNC:HGNC:1488|Ensembl:ENSG00000116489|HPRD:03347|Vega:OTTHUMG00000011769 |
| Gene type | protein-coding |
| Map location | 1p13.2 |
| Pascal p-value | 1.155E-4 |
| Sherlock p-value | 0.55 |
| Fetal beta | 0.154 |
| eGene | Cerebellum Myers' cis & trans Meta |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CAPZA1 | chr1 | 113209767 | G | A | NM_006135 NM_006135 | . . | silent splice | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1759687 | chr1 | 112951818 | CAPZA1 | 829 | 0.05 | cis | ||
| rs1175630 | chr1 | 112953340 | CAPZA1 | 829 | 0.09 | cis | ||
| rs1175645 | chr1 | 112981411 | CAPZA1 | 829 | 0.1 | cis | ||
| rs910697 | chr1 | 113063124 | CAPZA1 | 829 | 0.04 | cis | ||
| rs12401734 | chr1 | 113071833 | CAPZA1 | 829 | 1.207E-7 | cis | ||
| rs7551933 | chr1 | 113086705 | CAPZA1 | 829 | 2.362E-7 | cis | ||
| rs6681371 | chr1 | 113101167 | CAPZA1 | 829 | 1.207E-7 | cis | ||
| rs12406906 | chr1 | 113117899 | CAPZA1 | 829 | 1.126E-7 | cis | ||
| rs6666579 | chr1 | 113132392 | CAPZA1 | 829 | 0.14 | cis | ||
| rs2932537 | chr1 | 113230393 | CAPZA1 | 829 | 0 | cis | ||
| rs12401734 | chr1 | 113071833 | CAPZA1 | 829 | 2.646E-5 | trans | ||
| rs7551933 | chr1 | 113086705 | CAPZA1 | 829 | 4.955E-5 | trans | ||
| rs6681371 | chr1 | 113101167 | CAPZA1 | 829 | 2.646E-5 | trans | ||
| rs12406906 | chr1 | 113117899 | CAPZA1 | 829 | 2.444E-5 | trans | ||
| rs2932537 | chr1 | 113230393 | CAPZA1 | 829 | 0.19 | trans |
Section II. Transcriptome annotation
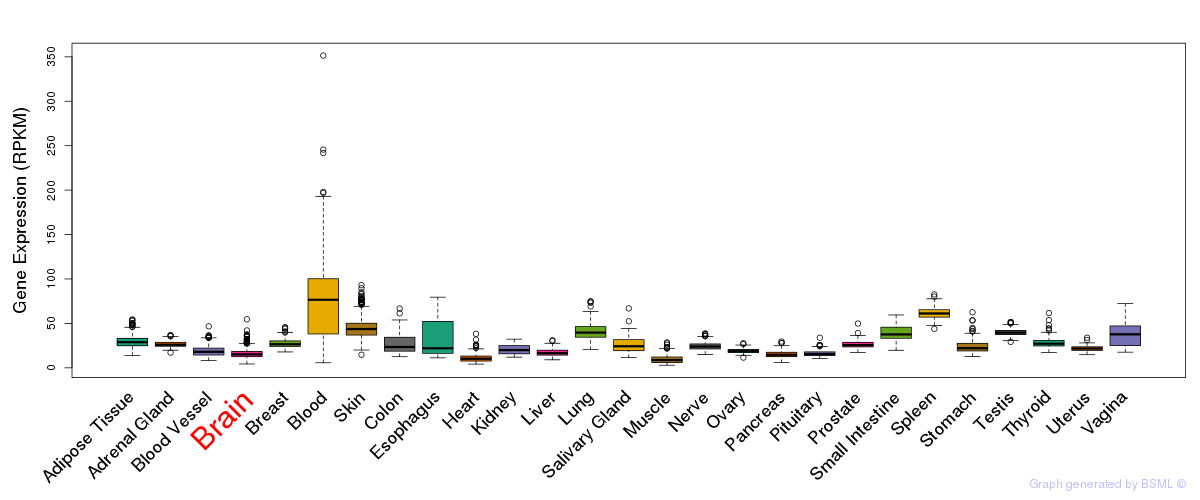
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GARS | 0.91 | 0.88 |
| PPP5C | 0.88 | 0.84 |
| HARS | 0.87 | 0.82 |
| SEC61A2 | 0.86 | 0.82 |
| PPP2R5D | 0.86 | 0.81 |
| PSMD7 | 0.86 | 0.83 |
| VDAC3 | 0.86 | 0.84 |
| C14orf153 | 0.85 | 0.84 |
| RAP1GDS1 | 0.84 | 0.82 |
| PSMD1 | 0.84 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.2 | -0.76 | -0.69 |
| AF347015.33 | -0.75 | -0.67 |
| AF347015.31 | -0.75 | -0.67 |
| AF347015.8 | -0.75 | -0.66 |
| MT-CO2 | -0.75 | -0.65 |
| AF347015.21 | -0.74 | -0.64 |
| AF347015.27 | -0.73 | -0.67 |
| MT-CYB | -0.72 | -0.64 |
| AF347015.15 | -0.71 | -0.64 |
| AF347015.26 | -0.71 | -0.64 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003779 | actin binding | TAS | 7665558 | |
| GO:0005515 | protein binding | IPI | 17353931 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006461 | protein complex assembly | TAS | 9331217 | |
| GO:0006928 | cell motion | TAS | 16130169 | |
| GO:0030036 | actin cytoskeleton organization | IEA | - | |
| GO:0051016 | barbed-end actin filament capping | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008290 | F-actin capping protein complex | TAS | 9331217 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | 132 | 101 | All SZGR 2.0 genes in this pathway |
| REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| SEIDEN ONCOGENESIS BY MET | 88 | 53 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| KEEN RESPONSE TO ROSIGLITAZONE DN | 106 | 68 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 VIA ERCC6 UP | 40 | 30 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN | 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN DN | 88 | 68 | All SZGR 2.0 genes in this pathway |
| KRIEG KDM3A TARGETS NOT HYPOXIA | 208 | 107 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 1073 | 1079 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-140 | 633 | 639 | 1A | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-144 | 137 | 143 | m8 | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-186 | 165 | 171 | m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-205 | 823 | 829 | 1A | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-214 | 773 | 779 | 1A | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-26 | 628 | 634 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-27 | 1073 | 1079 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-30-5p | 1185 | 1192 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-330 | 501 | 508 | 1A,m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-369-3p | 1396 | 1402 | m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-376 | 1195 | 1201 | m8 | hsa-miR-376a | AUCAUAGAGGAAAAUCCACGU |
| hsa-miR-376b | AUCAUAGAGGAAAAUCCAUGUU | ||||
| miR-485-5p | 126 | 132 | 1A | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
| miR-495 | 1333 | 1339 | m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-505 | 141 | 147 | 1A | hsa-miR-505 | GUCAACACUUGCUGGUUUCCUC |
| miR-7 | 1245 | 1251 | m8 | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
| miR-9 | 928 | 934 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.