Gene Page: CASP4
Summary ?
| GeneID | 837 |
| Symbol | CASP4 |
| Synonyms | ICE(rel)II|ICEREL-II|ICH-2|Mih1/TX|TX |
| Description | caspase 4 |
| Reference | MIM:602664|HGNC:HGNC:1505|Ensembl:ENSG00000196954|HPRD:04047|Vega:OTTHUMG00000166078 |
| Gene type | protein-coding |
| Map location | 11q22.2-q22.3 |
| Pascal p-value | 0.021 |
| Fetal beta | -0.973 |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Girard_2011 | Whole Exome Sequencing analysis | The data set included 15 DNMs found from the exomes of 14 schizophrenia probands and their parents. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 1.6047 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CASP4 | chr11 | 104825480 | T | C | NM_001225 NM_033306 | p.86K>E p.30K>E | missense missense | Schizophrenia | DNM:Girard_2011 |
Section II. Transcriptome annotation
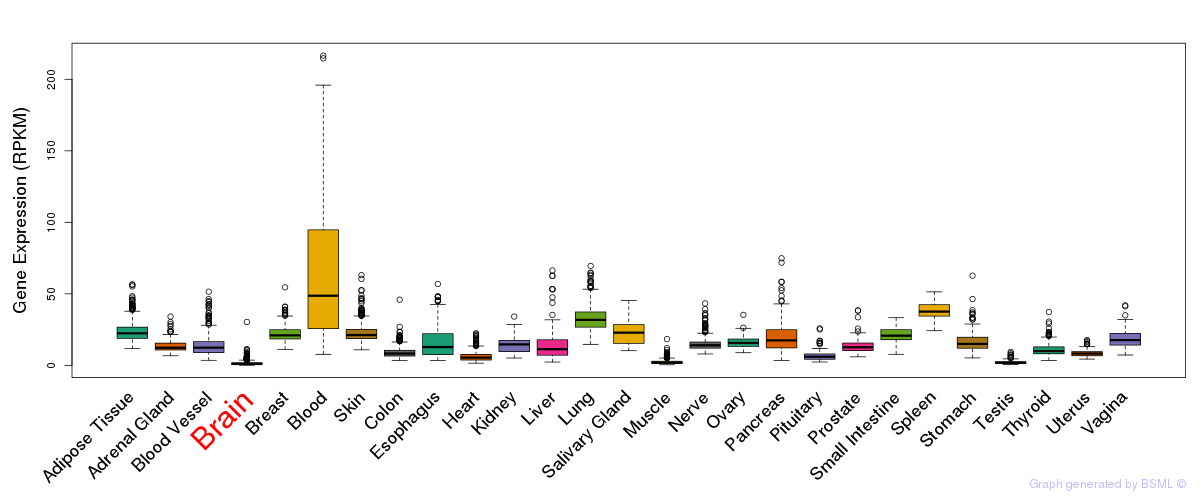
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DNAH7 | 0.61 | 0.49 |
| SPAG1 | 0.57 | 0.42 |
| DNAH12 | 0.55 | 0.40 |
| KIAA1529 | 0.55 | 0.41 |
| DNAH2 | 0.55 | 0.42 |
| CCDC65 | 0.54 | 0.41 |
| DNAH6 | 0.54 | 0.39 |
| WDR66 | 0.54 | 0.41 |
| FAM154B | 0.54 | 0.39 |
| WDR52 | 0.54 | 0.40 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TAF10 | -0.29 | -0.37 |
| FAM128A | -0.28 | -0.36 |
| NRL | -0.28 | -0.27 |
| AF347015.21 | -0.27 | -0.19 |
| UQCR | -0.27 | -0.34 |
| SERF2 | -0.26 | -0.30 |
| FAM128B | -0.26 | -0.31 |
| C22orf32 | -0.25 | -0.29 |
| COX6A1 | -0.25 | -0.25 |
| C19orf53 | -0.25 | -0.28 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 17353931 | |
| GO:0004197 | cysteine-type endopeptidase activity | IEA | - | |
| GO:0008233 | peptidase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006508 | proteolysis | IEA | - | |
| GO:0006508 | proteolysis | TAS | 7797510 | |
| GO:0006917 | induction of apoptosis | TAS | 7743998 | |
| GO:0042981 | regulation of apoptosis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APAF1 | CED4 | DKFZp781B1145 | apoptotic peptidase activating factor 1 | - | HPRD,BioGRID | 9539746 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | - | HPRD | 15123740 |
| CASP1 | ICE | IL1BC | P45 | caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | Reconstituted Complex | BioGRID | 10477277 |
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 8962078 |
| CASP14 | MGC119078 | MGC119079 | caspase 14, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 9792675 |
| CASP4 | ICE(rel)II | ICEREL-II | ICH-2 | Mih1/TX | TX | caspase 4, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 10872455 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | Biochemical Activity | BioGRID | 8962078 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | Affinity Capture-MS | BioGRID | 17353931 |
| CTNNBIP1 | ICAT | MGC15093 | catenin, beta interacting protein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| DSP | DPI | DPII | desmoplakin | - | HPRD | 15500642 |
| EIF2S3 | EIF2 | EIF2G | EIF2gamma | eIF-2gA | eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa | Affinity Capture-MS | BioGRID | 17353931 |
| GCN1L1 | GCN1 | GCN1L | KIAA0219 | GCN1 general control of amino-acid synthesis 1-like 1 (yeast) | Affinity Capture-MS | BioGRID | 17353931 |
| GSN | DKFZp313L0718 | gelsolin (amyloidosis, Finnish type) | Affinity Capture-MS | BioGRID | 17353931 |
| HEATR2 | FLJ20397 | FLJ25564 | FLJ31671 | FLJ39381 | HEAT repeat containing 2 | Affinity Capture-MS | BioGRID | 17353931 |
| IL18 | IGIF | IL-18 | IL-1g | IL1F4 | MGC12320 | interleukin 18 (interferon-gamma-inducing factor) | Biochemical Activity | BioGRID | 12096920 |
| IL1F7 | FIL1 | FIL1(ZETA) | FIL1Z | IL-1F7 | IL-1H4 | IL-1RP1 | IL1H4 | IL1RP1 | interleukin 1 family, member 7 (zeta) | Biochemical Activity | BioGRID | 12096920 |
| KIAA0776 | RP3-393D12.1 | KIAA0776 | Affinity Capture-MS | BioGRID | 17353931 |
| MSH6 | GTBP | HNPCC5 | HSAP | mutS homolog 6 (E. coli) | Affinity Capture-MS | BioGRID | 17353931 |
| MTHFD1L | DKFZp586G1517 | FLJ21145 | FTHFSDC1 | MTC1THFS | dJ292B18.2 | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like | Affinity Capture-MS | BioGRID | 17353931 |
| MYCBPAP | AMAP-1 | AMAP1 | DKFZp434N1415 | MYCBP associated protein | Affinity Capture-MS | BioGRID | 17353931 |
| NOD1 | CARD4 | CLR7.1 | NLRC1 | nucleotide-binding oligomerization domain containing 1 | - | HPRD,BioGRID | 10329646 |
| NUP93 | KIAA0095 | MGC21106 | nucleoporin 93kDa | Affinity Capture-MS | BioGRID | 17353931 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | PS1 interacts with caspase 4. This interaction was modeled on a demonstrated interaction between human PS1 and mouse caspase 4. | BIND | 10069390 |
| PSMB1 | FLJ25321 | HC5 | KIAA1838 | PMSB1 | PSC5 | proteasome (prosome, macropain) subunit, beta type, 1 | Affinity Capture-MS | BioGRID | 17353931 |
| SERPINB9 | CAP-3 | CAP3 | PI9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 | - | HPRD | 10477277 |
| SLC25A22 | FLJ13044 | GC1 | solute carrier family 25 (mitochondrial carrier: glutamate), member 22 | Affinity Capture-MS | BioGRID | 17353931 |
| SMC1A | CDLS2 | DKFZp686L19178 | DXS423E | KIAA0178 | MGC138332 | SB1.8 | SMC1 | SMC1L1 | SMC1alpha | SMCB | structural maintenance of chromosomes 1A | Affinity Capture-MS | BioGRID | 17353931 |
| SMC2 | CAP-E | CAPE | FLJ10093 | SMC2L1 | hCAP-E | structural maintenance of chromosomes 2 | Affinity Capture-MS | BioGRID | 17353931 |
| SMC3 | BAM | BMH | CDLS3 | CSPG6 | HCAP | SMC3L1 | structural maintenance of chromosomes 3 | Affinity Capture-MS | BioGRID | 17353931 |
| SRPRB | APMCF1 | signal recognition particle receptor, B subunit | Affinity Capture-MS | BioGRID | 17353931 |
| USP9X | DFFRX | FAF | FAM | ubiquitin specific peptidase 9, X-linked | Affinity Capture-MS | BioGRID | 17353931 |
| VDAC3 | HD-VDAC3 | voltage-dependent anion channel 3 | Affinity Capture-MS | BioGRID | 17353931 |
| XIAP | API3 | BIRC4 | ILP1 | MIHA | XLP2 | X-linked inhibitor of apoptosis | Affinity Capture-Western | BioGRID | 11927604 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA CASPASE PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME NOD1 2 SIGNALING PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | 46 | 33 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE DN | 92 | 51 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS UP | 126 | 72 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 UP | 120 | 73 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP | 215 | 137 | All SZGR 2.0 genes in this pathway |
| MANN RESPONSE TO AMIFOSTINE UP | 20 | 12 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| ALCALA APOPTOSIS | 88 | 60 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| MANTOVANI NFKB TARGETS UP | 43 | 33 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE UP | 167 | 92 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| BASSO B LYMPHOCYTE NETWORK | 143 | 96 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 UP | 62 | 35 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS UP | 88 | 64 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 3 UP | 18 | 11 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR UP | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| ZHANG PROLIFERATING VS QUIESCENT | 51 | 41 | All SZGR 2.0 genes in this pathway |
| GAJATE RESPONSE TO TRABECTEDIN UP | 67 | 45 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 1 UP | 16 | 10 | All SZGR 2.0 genes in this pathway |
| SAFFORD T LYMPHOCYTE ANERGY | 87 | 54 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 2 UP | 23 | 13 | All SZGR 2.0 genes in this pathway |
| HARRIS BRAIN CANCER PROGENITORS | 44 | 23 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P2 | 79 | 55 | All SZGR 2.0 genes in this pathway |
| GALI TP53 TARGETS APOPTOTIC UP | 8 | 7 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN | 252 | 155 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS UP | 77 | 56 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF LCP WITH H3K4ME3 | 128 | 68 | All SZGR 2.0 genes in this pathway |
| ONO AML1 TARGETS DN | 41 | 25 | All SZGR 2.0 genes in this pathway |
| ONO FOXP3 TARGETS DN | 42 | 23 | All SZGR 2.0 genes in this pathway |
| KESHELAVA MULTIPLE DRUG RESISTANCE | 88 | 56 | All SZGR 2.0 genes in this pathway |
| FONTAINE FOLLICULAR THYROID ADENOMA DN | 68 | 45 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA LB DN | 44 | 23 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 0 | 76 | 54 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN | 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN DN | 88 | 68 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| KIM GLIS2 TARGETS UP | 84 | 61 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| ALTEMEIER RESPONSE TO LPS WITH MECHANICAL VENTILATION | 128 | 81 | All SZGR 2.0 genes in this pathway |