Gene Page: CASP10
Summary ?
| GeneID | 843 |
| Symbol | CASP10 |
| Synonyms | ALPS2|FLICE2|MCH4 |
| Description | caspase 10 |
| Reference | MIM:601762|HGNC:HGNC:1500|Ensembl:ENSG00000003400|HPRD:03458|Vega:OTTHUMG00000132818 |
| Gene type | protein-coding |
| Map location | 2q33-q34 |
| Pascal p-value | 0.11 |
| Sherlock p-value | 0.247 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04152675 | 2 | 202085364 | CASP10 | 8.03E-5 | 0.266 | 0.025 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
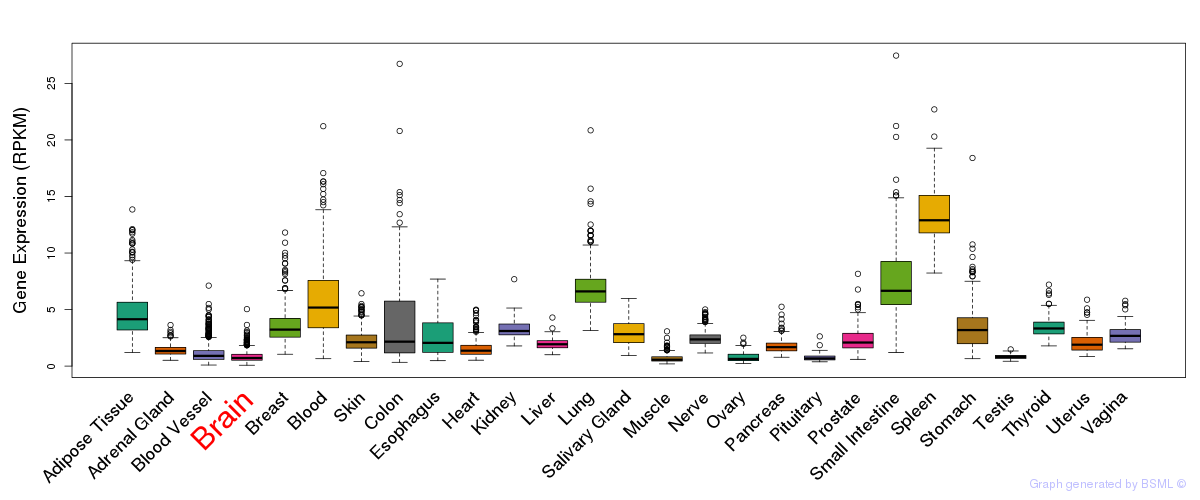
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SEPT2 | 0.79 | 0.77 |
| FBXO8 | 0.79 | 0.72 |
| ALDH9A1 | 0.78 | 0.78 |
| ELP4 | 0.77 | 0.70 |
| P4HA1 | 0.76 | 0.65 |
| PSAT1 | 0.76 | 0.70 |
| PTTG1IP | 0.75 | 0.70 |
| CDC14A | 0.74 | 0.65 |
| PLOD2 | 0.72 | 0.62 |
| ABHD4 | 0.72 | 0.68 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM133A | -0.31 | -0.27 |
| RP9P | -0.29 | -0.25 |
| FRMPD4 | -0.29 | -0.21 |
| TMEM63C | -0.27 | -0.22 |
| CACNA1I | -0.26 | -0.20 |
| RTN4RL1 | -0.26 | -0.19 |
| STRCP | -0.26 | -0.21 |
| SYT7 | -0.26 | -0.18 |
| GABBR2 | -0.25 | -0.19 |
| FAM159B | -0.25 | -0.07 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARHGDIA | GDIA1 | MGC117248 | RHOGDI | RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha | Co-purification | BioGRID | 15659383 |
| BFAR | BAR | RNF47 | bifunctional apoptosis regulator | - | HPRD | 10716992 |
| BID | FP497 | MGC15319 | MGC42355 | BH3 interacting domain death agonist | Co-purification | BioGRID | 15659383 |
| CASP1 | ICE | IL1BC | P45 | caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | Biochemical Activity | BioGRID | 8962078 |
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | Biochemical Activity | BioGRID | 8962078 |
| CASP14 | MGC119078 | MGC119079 | caspase 14, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 9792675 |
| CASP2 | CASP-2 | ICH-1L | ICH-1L/1S | ICH1 | NEDD2 | caspase 2, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 8962078 |
| CASP3 | CPP32 | CPP32B | SCA-1 | caspase 3, apoptosis-related cysteine peptidase | Biochemical Activity | BioGRID | 8962078 |
| CASP4 | ICE(rel)II | ICEREL-II | ICH-2 | Mih1/TX | TX | caspase 4, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 8962078 |
| CASP6 | MCH2 | caspase 6, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 8962078 |
| CASP7 | CMH-1 | ICE-LAP3 | MCH3 | caspase 7, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 8962078 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | Affinity Capture-Western Biochemical Activity Co-purification | BioGRID | 8962078 |11832478 |12887920 |15659383 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | - | HPRD | 9325248 |
| CASP9 | APAF-3 | APAF3 | CASPASE-9c | ICE-LAP6 | MCH6 | caspase 9, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 8962078 |
| CCAR1 | CARP-1 | CARP1 | MGC44628 | RP11-437A18.1 | cell division cycle and apoptosis regulator 1 | - | HPRD | 15069192 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | - | HPRD | 9208847 |9228018 |9289491 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9228018 |9289491 |12887920 |
| DEDD | CASP8IP1 | DEDD1 | DEFT | FLDED1 | KE05 | death effector domain containing | - | HPRD,BioGRID | 12527898 |
| DEDD2 | FLAME-3 | death effector domain containing 2 | - | HPRD,BioGRID | 12527898 |
| DSP | DPI | DPII | desmoplakin | - | HPRD | 15500642 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | Co-purification | BioGRID | 15659383 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | - | HPRD | 9045686 |11717445|11717445 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | - | HPRD,BioGRID | 11717445 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | - | HPRD,BioGRID | 9045686 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | Co-purification | BioGRID | 15659383 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | Affinity Capture-Western | BioGRID | 11002417 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | Co-purification | BioGRID | 15659383 |
| MSN | - | moesin | Co-purification | BioGRID | 15659383 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | CASP10 (caspase-10) cleaves RB-1 (hRB) at the DSID consensus site to produce the p68 form. | BIND | 15735701 |
| RHOA | ARH12 | ARHA | RHO12 | RHOH12 | ras homolog gene family, member A | Co-purification | BioGRID | 15659383 |
| RIPK1 | FLJ39204 | RIP | RIP1 | receptor (TNFRSF)-interacting serine-threonine kinase 1 | Affinity Capture-Western | BioGRID | 11002417 |
| RYBP | AAP1 | DEDAF | YEAF1 | RING1 and YY1 binding protein | - | HPRD,BioGRID | 11395500 |
| TNFRSF10A | APO2 | CD261 | DR4 | MGC9365 | TRAILR-1 | TRAILR1 | tumor necrosis factor receptor superfamily, member 10a | Affinity Capture-Western | BioGRID | 9325248 |
| TNFRSF10B | CD262 | DR5 | KILLER | KILLER/DR5 | TRAIL-R2 | TRAILR2 | TRICK2 | TRICK2A | TRICK2B | TRICKB | ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b | Affinity Capture-Western Co-purification | BioGRID | 9325248 |15659383 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | - | HPRD | 11098060 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | Affinity Capture-Western Co-purification | BioGRID | 9045686 |15659383 |
| TRAF1 | EBI6 | MGC:10353 | TNF receptor-associated factor 1 | Biochemical Activity | BioGRID | 11098060 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CASPASE PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA D4GDI PATHWAY | 13 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FAS PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DEATH PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| SA CASPASE CASCADE | 19 | 13 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID TRAIL PATHWAY | 28 | 20 | All SZGR 2.0 genes in this pathway |
| PID FAS PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | 13 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER UP | 206 | 111 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA DN | 104 | 59 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| NUNODA RESPONSE TO DASATINIB IMATINIB DN | 13 | 10 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP | 207 | 128 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| CALVET IRINOTECAN SENSITIVE VS RESISTANT DN | 5 | 5 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE UP | 78 | 51 | All SZGR 2.0 genes in this pathway |
| HOFMANN CELL LYMPHOMA DN | 39 | 29 | All SZGR 2.0 genes in this pathway |
| CHIARETTI ACUTE LYMPHOBLASTIC LEUKEMIA ZAP70 | 67 | 33 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR UP | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C3 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| ZHAN EARLY DIFFERENTIATION GENES UP | 7 | 6 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 1HR UP | 17 | 10 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |