Gene Page: FUT10
Summary ?
| GeneID | 84750 |
| Symbol | FUT10 |
| Synonyms | - |
| Description | fucosyltransferase 10 (alpha (1,3) fucosyltransferase) |
| Reference | HGNC:HGNC:19234|Ensembl:ENSG00000172728|Vega:OTTHUMG00000163954 |
| Gene type | protein-coding |
| Map location | 8p12 |
| Pascal p-value | 0.612 |
| Fetal beta | -0.191 |
| DMG | 1 (# studies) |
| eGene | Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04687486 | 8 | 33330940 | FUT10 | 3.733E-4 | 0.393 | 0.043 | DMG:Wockner_2014 |
| cg07391088 | 8 | 33330912 | FUT10 | 3.84E-4 | 0.477 | 0.043 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
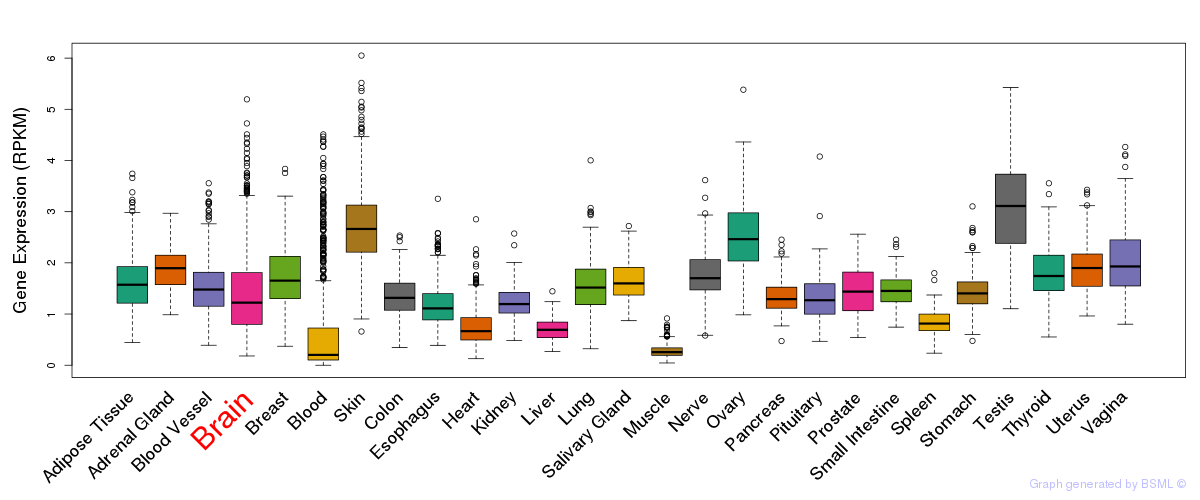
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0016757 | transferase activity, transferring glycosyl groups | IEA | - | |
| GO:0046920 | alpha(1,3)-fucosyltransferase activity | TAS | 11698403 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | NAS | neurite (GO term level: 5) | 11698403 |
| GO:0006457 | protein folding | NAS | 11698403 | |
| GO:0006486 | protein amino acid glycosylation | TAS | 11698403 | |
| GO:0009790 | embryonic development | NAS | 11698403 | |
| GO:0009566 | fertilization | NAS | 11698403 | |
| GO:0006605 | protein targeting | NAS | 11698403 | |
| GO:0042060 | wound healing | NAS | 11698403 | |
| GO:0042355 | L-fucose catabolic process | NAS | 11698403 | |
| GO:0030097 | hemopoiesis | NAS | 11698403 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | TAS | 11698403 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| WAGNER APO2 SENSITIVITY | 25 | 14 | All SZGR 2.0 genes in this pathway |