Gene Page: HIRIP3
Summary ?
| GeneID | 8479 |
| Symbol | HIRIP3 |
| Synonyms | - |
| Description | HIRA interacting protein 3 |
| Reference | MIM:603365|HGNC:HGNC:4917|Ensembl:ENSG00000149929|HPRD:04531|Vega:OTTHUMG00000132118 |
| Gene type | protein-coding |
| Map location | 16p11.2 |
| Pascal p-value | 2.515E-9 |
| Sherlock p-value | 0.081 |
| Fetal beta | 0.141 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01775 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05995267 | 16 | 29822529 | HIRIP3 | 7.949E-4 | 4.713 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
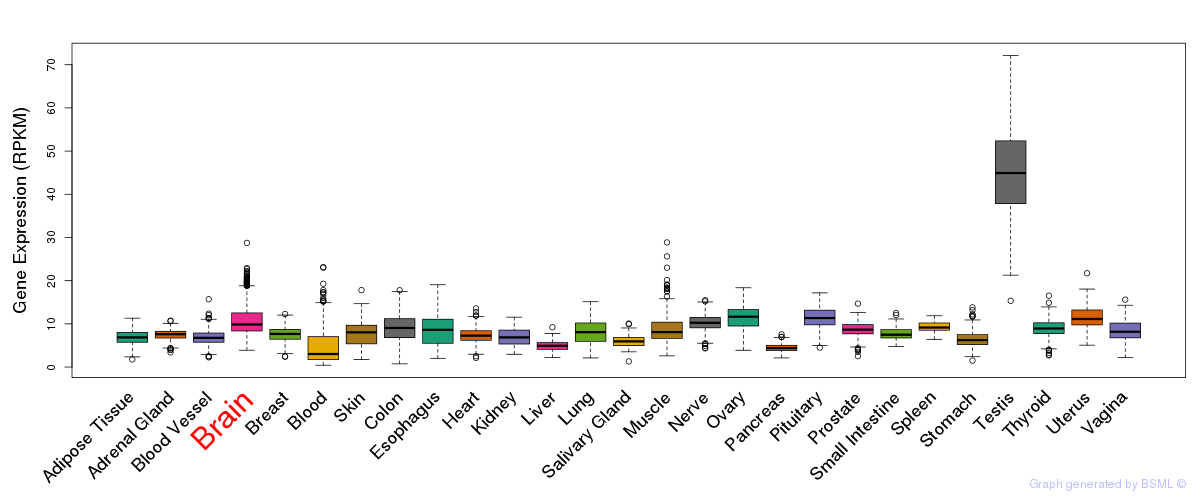
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KBTBD7 | 0.97 | 0.97 |
| SCYL2 | 0.96 | 0.97 |
| WDR82 | 0.96 | 0.96 |
| KBTBD6 | 0.96 | 0.96 |
| RBMXL1 | 0.96 | 0.95 |
| ASB7 | 0.95 | 0.95 |
| ZMYM4 | 0.95 | 0.97 |
| SPAST | 0.95 | 0.96 |
| GRPEL2 | 0.95 | 0.95 |
| C5orf24 | 0.95 | 0.96 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.74 | -0.87 |
| MT-CO2 | -0.73 | -0.87 |
| AF347015.31 | -0.72 | -0.85 |
| AIFM3 | -0.72 | -0.77 |
| AF347015.33 | -0.72 | -0.84 |
| AF347015.27 | -0.72 | -0.84 |
| AF347015.8 | -0.71 | -0.87 |
| HSD17B14 | -0.71 | -0.79 |
| S100B | -0.71 | -0.81 |
| MT-CYB | -0.71 | -0.84 |
Section III. Gene Ontology annotation
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0006333 | chromatin assembly or disassembly | TAS | 9710638 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 9710638 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| JIANG VHL TARGETS | 138 | 91 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL SHORT TERM | 32 | 15 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL UP | 45 | 26 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA VIA VHL | 34 | 24 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |