Gene Page: LSM10
Summary ?
| GeneID | 84967 |
| Symbol | LSM10 |
| Synonyms | MST074|MSTP074 |
| Description | LSM10, U7 small nuclear RNA associated |
| Reference | HGNC:HGNC:17562|Ensembl:ENSG00000181817|HPRD:14326|Vega:OTTHUMG00000008140 |
| Gene type | protein-coding |
| Map location | 1p34.3 |
| Pascal p-value | 0.492 |
| Sherlock p-value | 0.279 |
| Fetal beta | -0.583 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0192 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg00648152 | 1 | 36863595 | LSM10 | 5.014E-4 | -0.276 | 0.047 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
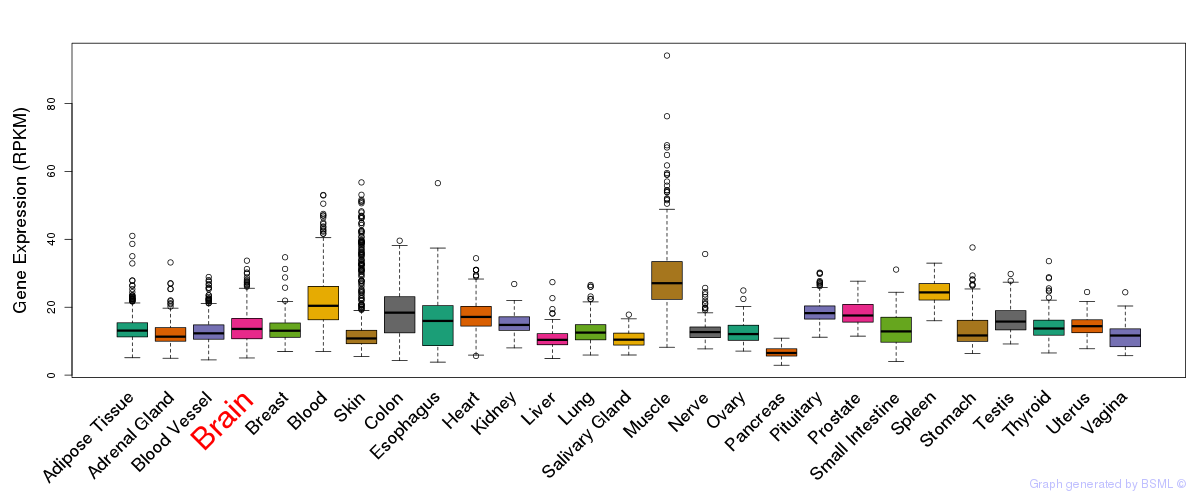
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003723 | RNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 15231747 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006397 | mRNA processing | IEA | - | |
| GO:0008380 | RNA splicing | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0030529 | ribonucleoprotein complex | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | 11 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION | 105 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | 23 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | 44 | 22 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO CYCLOPHOSPHAMIDE | 18 | 12 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS DN | 187 | 109 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| BOUDOUKHA BOUND BY IGF2BP2 | 111 | 59 | All SZGR 2.0 genes in this pathway |