Gene Page: ITGA8
Summary ?
| GeneID | 8516 |
| Symbol | ITGA8 |
| Synonyms | - |
| Description | integrin subunit alpha 8 |
| Reference | MIM:604063|HGNC:HGNC:6144|Ensembl:ENSG00000077943|HPRD:10361|Vega:OTTHUMG00000017733 |
| Gene type | protein-coding |
| Map location | 10p13 |
| Pascal p-value | 0.489 |
| Fetal beta | -0.682 |
| eGene | Caudate basal ganglia Myers' cis & trans Meta |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7370564 | 0 | ITGA8 | 8516 | 0.07 | trans |
Section II. Transcriptome annotation
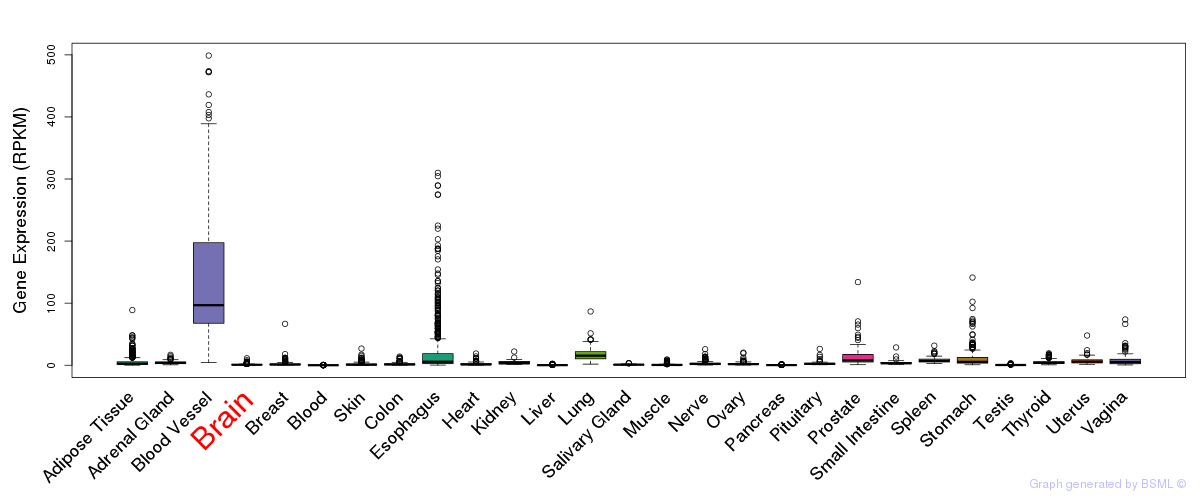
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030030 | cell projection organization | IEA | axon (GO term level: 7) | - |
| GO:0007399 | nervous system development | IEA | neurite (GO term level: 5) | - |
| GO:0001656 | metanephros development | IEA | - | |
| GO:0007160 | cell-matrix adhesion | NAS | 7768999 | |
| GO:0007229 | integrin-mediated signaling pathway | IEA | - | |
| GO:0007613 | memory | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0016337 | cell-cell adhesion | NAS | 7768999 | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0042472 | inner ear morphogenesis | IEA | - | |
| GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | IEA | - | |
| GO:0030198 | extracellular matrix organization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0008305 | integrin complex | TAS | 7768999 | |
| GO:0045177 | apical part of cell | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ECM RECEPTOR INTERACTION | 84 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN1 PATHWAY | 66 | 44 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN CS PATHWAY | 26 | 16 | All SZGR 2.0 genes in this pathway |
| PID ARF6 TRAFFICKING PATHWAY | 49 | 34 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| HAEGERSTRAND RESPONSE TO IMATINIB | 9 | 5 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP | 176 | 111 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL UP | 120 | 89 | All SZGR 2.0 genes in this pathway |
| TUOMISTO TUMOR SUPPRESSION BY COL13A1 UP | 18 | 9 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| DAVIES MULTIPLE MYELOMA VS MGUS UP | 13 | 10 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT UP | 166 | 105 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |