Gene Page: CAV1
Summary ?
| GeneID | 857 |
| Symbol | CAV1 |
| Synonyms | BSCL3|CGL3|LCCNS|MSTP085|PPH3|VIP21 |
| Description | caveolin 1 |
| Reference | MIM:601047|HGNC:HGNC:1527|Ensembl:ENSG00000105974|HPRD:03028|Vega:OTTHUMG00000023413 |
| Gene type | protein-coding |
| Map location | 7q31.1 |
| Pascal p-value | 0.592 |
| Fetal beta | 0.013 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.5838 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
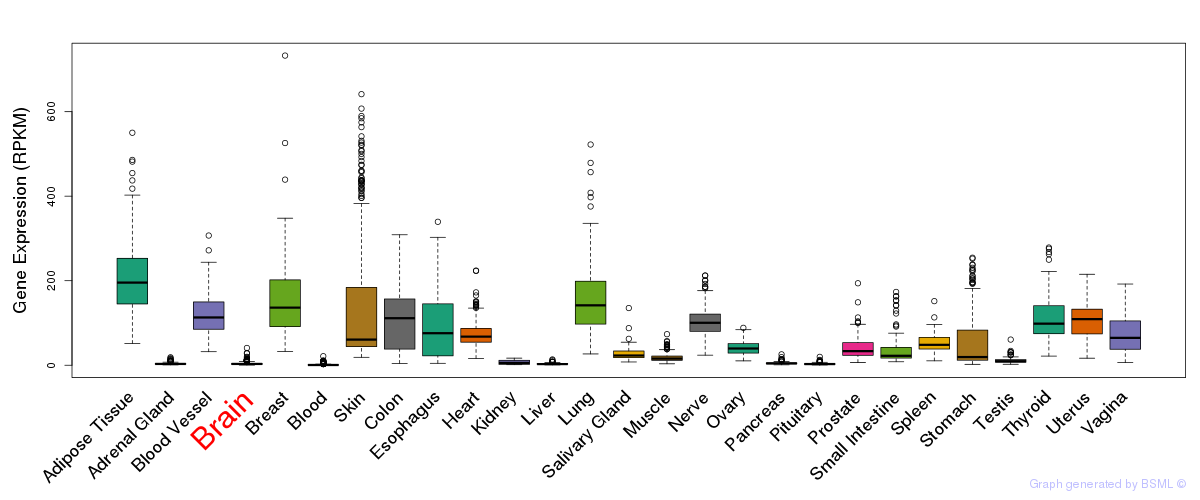
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IL17RA | 0.67 | 0.45 |
| NFATC2 | 0.64 | 0.55 |
| LINGO3 | 0.61 | 0.29 |
| ADCY5 | 0.60 | 0.43 |
| OTOF | 0.60 | 0.39 |
| ITGB2 | 0.60 | 0.67 |
| PMEPA1 | 0.59 | 0.43 |
| SH3D20 | 0.59 | 0.42 |
| ENTPD3 | 0.59 | 0.36 |
| GPR52 | 0.59 | 0.34 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AL139819.3 | -0.28 | -0.35 |
| RP9P | -0.27 | -0.39 |
| AC120053.1 | -0.24 | -0.30 |
| SYCP3 | -0.24 | -0.32 |
| AC010300.1 | -0.24 | -0.29 |
| NOX1 | -0.23 | -0.25 |
| AC087071.1 | -0.22 | -0.20 |
| FAM159B | -0.22 | -0.37 |
| AF347015.21 | -0.22 | -0.23 |
| SPDYA | -0.22 | -0.27 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 16951145 | |
| GO:0005198 | structural molecule activity | IDA | 11865038 | |
| GO:0016504 | protease activator activity | ISS | - | |
| GO:0015485 | cholesterol binding | TAS | 11294831 | |
| GO:0050998 | nitric-oxide synthase binding | IPI | 12852865 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007519 | skeletal muscle development | ISS | neuron (GO term level: 8) | - |
| GO:0000188 | inactivation of MAPK activity | ISS | - | |
| GO:0001570 | vasculogenesis | ISS | - | |
| GO:0001666 | response to hypoxia | ISS | - | |
| GO:0001937 | negative regulation of endothelial cell proliferation | ISS | - | |
| GO:0016050 | vesicle organization | IDA | 12743374 | |
| GO:0052547 | regulation of peptidase activity | ISS | - | |
| GO:0008104 | protein localization | ISS | - | |
| GO:0019915 | sequestering of lipid | ISS | - | |
| GO:0006816 | calcium ion transport | ISS | - | |
| GO:0006940 | regulation of smooth muscle contraction | ISS | - | |
| GO:0006641 | triacylglycerol metabolic process | ISS | - | |
| GO:0006897 | endocytosis | ISS | - | |
| GO:0006874 | cellular calcium ion homeostasis | ISS | - | |
| GO:0030193 | regulation of blood coagulation | IMP | 17848177 | |
| GO:0033484 | nitric oxide homeostasis | ISS | - | |
| GO:0051260 | protein homooligomerization | ISS | - | |
| GO:0030857 | negative regulation of epithelial cell differentiation | ISS | - | |
| GO:0019217 | regulation of fatty acid metabolic process | ISS | - | |
| GO:0030301 | cholesterol transport | TAS | 11294831 | |
| GO:0043409 | negative regulation of MAPKKK cascade | ISS | - | |
| GO:0042632 | cholesterol homeostasis | TAS | 11294831 | |
| GO:0045907 | positive regulation of vasoconstriction | ISS | - | |
| GO:0045908 | negative regulation of vasodilation | ISS | - | |
| GO:0045019 | negative regulation of nitric oxide biosynthetic process | ISS | - | |
| GO:0046426 | negative regulation of JAK-STAT cascade | ISS | - | |
| GO:0048554 | positive regulation of metalloenzyme activity | ISS | - | |
| GO:0060056 | mammary gland involution | ISS | - | |
| GO:0051899 | membrane depolarization | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000139 | Golgi membrane | EXP | 14707126 | |
| GO:0000139 | Golgi membrane | IDA | 11294831 | |
| GO:0000139 | Golgi membrane | IEA | - | |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005811 | lipid particle | EXP | 9188442 |9580552 |10781589 |16722822 |16807357 | |
| GO:0005783 | endoplasmic reticulum | IDA | 11294831 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005901 | caveola | IDA | 15219854 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 1360410 | |
| GO:0048471 | perinuclear region of cytoplasm | ISS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADRBK1 | BARK1 | BETA-ARK1 | FLJ16718 | GRK2 | adrenergic, beta, receptor kinase 1 | Reconstituted Complex | BioGRID | 10085129 |
| AKAP1 | AKAP | AKAP121 | AKAP149 | AKAP84 | D-AKAP1 | MGC1807 | PRKA1 | SAKAP84 | A kinase (PRKA) anchor protein 1 | - | HPRD | 11546661 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | - | HPRD,BioGRID | 9553108 |
| AQP3 | GIL | aquaporin 3 (Gill blood group) | Affinity Capture-Western | BioGRID | 14675200 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 11278309 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Caveolin-1 interacts with AR. | BIND | 11278309 |
| BMX | ETK | PSCTK2 | PSCTK3 | BMX non-receptor tyrosine kinase | - | HPRD,BioGRID | 11751885 |
| BST1 | CD157 | bone marrow stromal cell antigen 1 | - | HPRD | 11866528 |
| BTK | AGMX1 | AT | ATK | BPK | IMD1 | MGC126261 | MGC126262 | PSCTK1 | XLA | Bruton agammaglobulinemia tyrosine kinase | - | HPRD,BioGRID | 11751885 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | CAV1 (caveolin-1) homodimerizes. | BIND | 15782218 |
| CAV2 | CAV | MGC12294 | caveolin 2 | - | HPRD,BioGRID | 9361015 |12414992 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | E-cadherin interacts with caveolin-1. | BIND | 14706341 |
| CSK | MGC117393 | c-src tyrosine kinase | - | HPRD,BioGRID | 11805080 |
| DAG1 | 156DAG | A3a | AGRNR | DAG | dystroglycan 1 (dystrophin-associated glycoprotein 1) | Reconstituted Complex | BioGRID | 11724572 |
| EDNRB | ABCDS | ETB | ETBR | ETRB | HSCR | HSCR2 | endothelin receptor type B | - | HPRD,BioGRID | 12694195 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 9374534 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | - | HPRD | 9685399 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD,BioGRID | 11563984 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | - | HPRD | 10637311 |
| FLOT1 | - | flotillin 1 | Affinity Capture-Western | BioGRID | 10212252 |
| FLOT2 | ECS-1 | ECS1 | ESA | ESA1 | M17S1 | flotillin 2 | - | HPRD,BioGRID | 10212252 |
| GJA1 | CX43 | DFNB38 | GJAL | ODDD | gap junction protein, alpha 1, 43kDa | - | HPRD,BioGRID | 11980479 |
| GJA3 | CX46 | CZP3 | gap junction protein, alpha 3, 46kDa | - | HPRD,BioGRID | 11980479 |
| GJB2 | CX26 | DFNA3 | DFNB1 | HID | KID | NSRD1 | PPK | gap junction protein, beta 2, 26kDa | - | HPRD,BioGRID | 11980479 |
| GRK1 | GPRK1 | RHOK | RK | G protein-coupled receptor kinase 1 | Reconstituted Complex | BioGRID | 10085129 |
| GRK5 | GPRK5 | G protein-coupled receptor kinase 5 | Reconstituted Complex | BioGRID | 10085129 |
| ILK | DKFZp686F1765 | P59 | integrin-linked kinase | ILK1 interacts with caveolin-1. | BIND | 15722199 |
| INSR | CD220 | HHF5 | insulin receptor | - | HPRD,BioGRID | 10598578 |
| MALL | BENE | MGC4419 | mal, T-cell differentiation protein-like | - | HPRD | 1294831 |
| NEU3 | FLJ12388 | SIAL3 | sialidase 3 (membrane sialidase) | - | HPRD,BioGRID | 12011038 |
| NGFR | CD271 | Gp80-LNGFR | TNFRSF16 | p75(NTR) | p75NTR | nerve growth factor receptor (TNFR superfamily, member 16) | Reconstituted Complex | BioGRID | 9867838 |
| NOS2 | HEP-NOS | INOS | NOS | NOS2A | nitric oxide synthase 2, inducible | - | HPRD,BioGRID | 11114180 |
| NOS3 | ECNOS | eNOS | nitric oxide synthase 3 (endothelial cell) | - | HPRD,BioGRID | 8910295 |
| NTRK1 | DKFZp781I14186 | MTC | TRK | TRK1 | TRKA | p140-TrkA | neurotrophic tyrosine kinase, receptor, type 1 | - | HPRD,BioGRID | 9867838 |
| PDGFRA | CD140A | MGC74795 | PDGFR2 | Rhe-PDGFRA | platelet-derived growth factor receptor, alpha polypeptide | Affinity Capture-Western Co-fractionation Reconstituted Complex | BioGRID | 10066366 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | Affinity Capture-Western Co-fractionation Reconstituted Complex | BioGRID | 10066366 |
| PLD2 | - | phospholipase D2 | - | HPRD,BioGRID | 10675563 |
| PPP1CA | MGC15877 | MGC1674 | PP-1A | PPP1A | protein phosphatase 1, catalytic subunit, alpha isoform | CAV1 interacts with PP1-C. | BIND | 14645548 |
| PRKACA | MGC102831 | MGC48865 | PKACA | protein kinase, cAMP-dependent, catalytic, alpha | Affinity Capture-Western | BioGRID | 11546661 |
| PRNP | ASCR | CD230 | CJD | GSS | MGC26679 | PRIP | PrP | PrP27-30 | PrP33-35C | PrPc | prion | prion protein | - | HPRD | 10988071 |
| PTEN | 10q23del | BZS | MGC11227 | MHAM | MMAC1 | PTEN1 | TEP1 | phosphatase and tensin homolog | - | HPRD,BioGRID | 12176037 |
| PTEN | 10q23del | BZS | MGC11227 | MHAM | MMAC1 | PTEN1 | TEP1 | phosphatase and tensin homolog | caveolin-1 interacts with PTEN. | BIND | 12176037 |
| PTGS2 | COX-2 | COX2 | GRIPGHS | PGG/HS | PGHS-2 | PHS-2 | hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | - | HPRD,BioGRID | 11432874 |
| PTPN1 | PTP1B | protein tyrosine phosphatase, non-receptor type 1 | caveolin-1 interacts with PTP1B. | BIND | 12176037 |
| PTPN1 | PTP1B | protein tyrosine phosphatase, non-receptor type 1 | - | HPRD,BioGRID | 12176037 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | - | HPRD,BioGRID | 12176037 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | caveolin-1 interacts with SH-PTP2. | BIND | 12176037 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | - | HPRD,BioGRID | 12176037 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | caveolin-1 interacts with PTP-1C. | BIND | 12176037 |
| PTPRF | FLJ43335 | FLJ45062 | FLJ45567 | LAR | protein tyrosine phosphatase, receptor type, F | - | HPRD,BioGRID | 12176037 |
| PTPRF | FLJ43335 | FLJ45062 | FLJ45567 | LAR | protein tyrosine phosphatase, receptor type, F | caveolin-1 interacts with LAR. | BIND | 12176037 |
| RCVRN | RCV1 | recoverin | - | HPRD,BioGRID | 12507501 |
| S1PR1 | CHEDG1 | D1S3362 | ECGF1 | EDG-1 | EDG1 | FLJ58121 | S1P1 | sphingosine-1-phosphate receptor 1 | - | HPRD,BioGRID | 10921915 |
| SCP2 | DKFZp686C12188 | DKFZp686D11188 | NLTP | NSL-TP | SCPX | sterol carrier protein 2 | Affinity Capture-Western FRET Two-hybrid | BioGRID | 15182174 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | Co-fractionation | BioGRID | 11102446 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 8910575 |
| STOML3 | Epb7.2l | SRO | stomatin (EPB72)-like 3 | - | HPRD | 12122055 |
| STRN | MGC125642 | SG2NA | striatin, calmodulin binding protein | - | HPRD | 11707266 |
| STRN4 | FLJ35594 | ZIN | zinedin | striatin, calmodulin binding protein 4 | - | HPRD | 11707266 |
| TGFBR1 | AAT5 | ACVRLK4 | ALK-5 | ALK5 | LDS1A | LDS2A | SKR4 | TGFR-1 | transforming growth factor, beta receptor 1 | - | HPRD,BioGRID | 11102446 |
| TNFRSF1B | CD120b | TBPII | TNF-R-II | TNF-R75 | TNFBR | TNFR1B | TNFR2 | TNFR80 | p75 | p75TNFR | tumor necrosis factor receptor superfamily, member 1B | - | HPRD,BioGRID | 11112773 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD,BioGRID | 11112773 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | - | HPRD,BioGRID | 11112773 |11805080 |
| TRPC1 | HTRP-1 | MGC133334 | MGC133335 | TRP1 | transient receptor potential cation channel, subfamily C, member 1 | Affinity Capture-Western | BioGRID | 10980191 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | Co-purification | BioGRID | 12147258 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG VIRAL MYOCARDITIS | 73 | 58 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NO1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTEGRIN PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| PID INSULIN PATHWAY | 45 | 32 | All SZGR 2.0 genes in this pathway |
| PID PTP1B PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID WNT CANONICAL PATHWAY | 20 | 18 | All SZGR 2.0 genes in this pathway |
| PID TNF PATHWAY | 46 | 33 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID ALK1 PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PID PDGFRA PATHWAY | 22 | 18 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 2 PATHWAY | 69 | 57 | All SZGR 2.0 genes in this pathway |
| PID TGFBR PATHWAY | 55 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | 91 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME ENOS ACTIVATION AND REGULATION | 20 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME BASIGIN INTERACTIONS | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | 46 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| PYEON HPV POSITIVE TUMORS DN | 10 | 5 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS LOBULAR NORMAL DN | 69 | 43 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 3D UP | 182 | 110 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D UP | 194 | 122 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| SCIBETTA KDM5B TARGETS DN | 81 | 55 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP | 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG UP | 130 | 85 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A TARGETS DN | 91 | 58 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A AND HIF2A TARGETS DN | 104 | 72 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER GRADES 1 2 DN | 67 | 43 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| CAIRO PML TARGETS BOUND BY MYC DN | 14 | 12 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| KERLEY RESPONSE TO CISPLATIN UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| MISSIAGLIA REGULATED BY METHYLATION DN | 122 | 67 | All SZGR 2.0 genes in this pathway |
| MIDORIKAWA AMPLIFIED IN LIVER CANCER | 55 | 38 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| WANG METHYLATED IN BREAST CANCER | 35 | 25 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER UP | 227 | 137 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| INAMURA LUNG CANCER SCC DN | 14 | 6 | All SZGR 2.0 genes in this pathway |
| WATTEL AUTONOMOUS THYROID ADENOMA DN | 55 | 29 | All SZGR 2.0 genes in this pathway |
| SASAI RESISTANCE TO NEOPLASTIC TRANSFROMATION | 50 | 31 | All SZGR 2.0 genes in this pathway |
| GROSS ELK3 TARGETS UP | 27 | 16 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN | 110 | 78 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| GUENTHER GROWTH SPHERICAL VS ADHERENT DN | 26 | 19 | All SZGR 2.0 genes in this pathway |
| BERTUCCI INVASIVE CARCINOMA DUCTAL VS LOBULAR DN | 46 | 34 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS MODERATELY DN | 110 | 64 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| ROSS ACUTE MYELOID LEUKEMIA CBF | 82 | 57 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE UP | 78 | 51 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH AML1 ETO FUSION | 76 | 55 | All SZGR 2.0 genes in this pathway |
| ZHANG TARGETS OF EWSR1 FLI1 FUSION | 88 | 68 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| CROMER METASTASIS DN | 81 | 58 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| LINDVALL IMMORTALIZED BY TERT UP | 78 | 48 | All SZGR 2.0 genes in this pathway |
| SIMBULAN PARP1 TARGETS UP | 31 | 23 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| MCDOWELL ACUTE LUNG INJURY DN | 48 | 33 | All SZGR 2.0 genes in this pathway |
| TRACEY RESISTANCE TO IFNA2 DN | 32 | 23 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| HARRIS BRAIN CANCER PROGENITORS | 44 | 23 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| SAGIV CD24 TARGETS UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| SAGIV CD24 TARGETS DN | 46 | 26 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY DN | 58 | 39 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS DN | 108 | 64 | All SZGR 2.0 genes in this pathway |
| HUANG DASATINIB RESISTANCE UP | 81 | 53 | All SZGR 2.0 genes in this pathway |
| MATTHEWS AP1 TARGETS | 17 | 12 | All SZGR 2.0 genes in this pathway |
| TAYLOR METHYLATED IN ACUTE LYMPHOBLASTIC LEUKEMIA | 77 | 52 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS DN | 142 | 79 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN MYELOMA VS MATURE B LYMPHOCYTE | 101 | 76 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN PLASMA CELL VS MATURE B LYMPHOCYTE | 67 | 51 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN ACTIVATED B LYMPHOCYTE | 81 | 66 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN ACTIVATED DENDRITIC CELL | 65 | 49 | All SZGR 2.0 genes in this pathway |
| ZHAN LATE DIFFERENTIATION GENES UP | 33 | 24 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 13 | 30 | 20 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| LI PROSTATE CANCER EPIGENETIC | 30 | 22 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE DN | 103 | 67 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| DELPUECH FOXO3 TARGETS UP | 68 | 49 | All SZGR 2.0 genes in this pathway |
| WIEDERSCHAIN TARGETS OF BMI1 AND PCGF2 | 57 | 39 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 655 | 661 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-142-5p | 1665 | 1671 | 1A | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-17-5p/20/93.mr/106/519.d | 1522 | 1528 | 1A | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-199 | 1573 | 1579 | m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-204/211 | 396 | 402 | 1A | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-30-3p | 1615 | 1622 | 1A,m8 | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-431 | 1727 | 1733 | 1A | hsa-miR-431 | UGUCUUGCAGGCCGUCAUGCA |
| miR-505 | 286 | 292 | 1A | hsa-miR-505 | GUCAACACUUGCUGGUUUCCUC |
| miR-96 | 55 | 61 | m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.