Gene Page: SOCS1
Summary ?
| GeneID | 8651 |
| Symbol | SOCS1 |
| Synonyms | CIS1|CISH1|JAB|SOCS-1|SSI-1|SSI1|TIP3 |
| Description | suppressor of cytokine signaling 1 |
| Reference | MIM:603597|HGNC:HGNC:19383|Ensembl:ENSG00000185338|HPRD:04669|Vega:OTTHUMG00000129792 |
| Gene type | protein-coding |
| Map location | 16p13.13 |
| Pascal p-value | 0.756 |
| Fetal beta | -0.294 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01717706 | 16 | 11349308 | SOCS1 | -0.026 | 0.69 | DMG:Nishioka_2013 | |
| cg26012103 | 16 | 11349372 | SOCS1 | -0.026 | 0.85 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
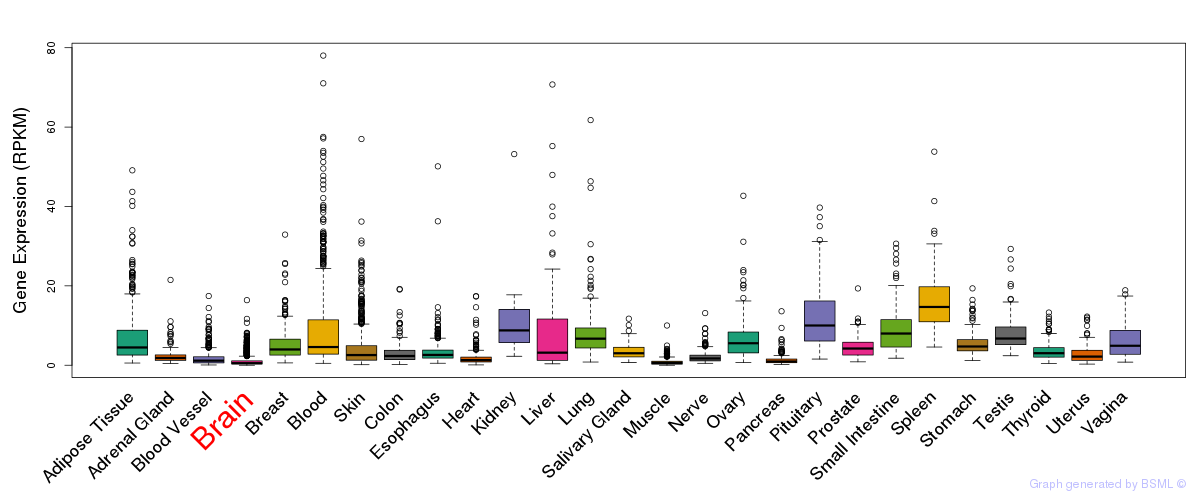
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ALDOA | 0.86 | 0.89 |
| TPST2 | 0.83 | 0.85 |
| P4HTM | 0.83 | 0.82 |
| RUNDC3A | 0.83 | 0.83 |
| TMX2 | 0.82 | 0.82 |
| ANXA7 | 0.82 | 0.84 |
| GNPTG | 0.82 | 0.84 |
| GBA | 0.81 | 0.83 |
| BDH1 | 0.81 | 0.81 |
| RHBDD2 | 0.81 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EIF5B | -0.57 | -0.64 |
| UPF3B | -0.54 | -0.59 |
| THOC2 | -0.53 | -0.54 |
| RBMX2 | -0.53 | -0.61 |
| KIAA1949 | -0.51 | -0.40 |
| FNBP1L | -0.51 | -0.35 |
| ZNF326 | -0.51 | -0.51 |
| TUBB2B | -0.50 | -0.46 |
| YBX1 | -0.50 | -0.42 |
| ATAD2B | -0.50 | -0.48 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AXL | JTK11 | UFO | AXL receptor tyrosine kinase | Axl interacts with SOCS-1. | BIND | 12470648 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | Reconstituted Complex | BioGRID | 10022833 |
| CSF1 | MCSF | MGC31930 | colony stimulating factor 1 (macrophage) | - | HPRD | 11297560 |
| CSF1R | C-FMS | CD115 | CSFR | FIM2 | FMS | colony stimulating factor 1 receptor | - | HPRD,BioGRID | 10022833 |11297560 |
| CSF1R | C-FMS | CD115 | CSFR | FIM2 | FMS | colony stimulating factor 1 receptor | Socs1 interacts with CSF1-R. This interaction was modeled on a demonstrated interaction between mouse proteins. | BIND | 10022833 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 12070153 |
| FLT3 | CD135 | FLK2 | STK1 | fms-related tyrosine kinase 3 | Socs1 interacts with Flt3. This interaction was modeled on a demonstrated interaction between mouse Socs1 and Flt3. | BIND | 10022833 |
| FLT3 | CD135 | FLK2 | STK1 | fms-related tyrosine kinase 3 | - | HPRD,BioGRID | 10022833 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | Socs1 interacts with Fyn. This interaction was modeled on a demonstrated interaction between mouse Socs1 and Fyn from an unspecified species. | BIND | 10022833 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 10022833 |
| GHR | GHBP | growth hormone receptor | - | HPRD,BioGRID | 10585430 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Socs1 interacts with Grb2. This interaction was modeled on a demonstrated interaction between mouse Socs1 and human Grb2. | BIND | 10022833 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 10022833 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | - | HPRD,BioGRID | 9727029 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | Insulin-like Growth Factor-I Receptor interacts with SOCS1 | BIND | 9727029 |
| IL2RB | CD122 | P70-75 | interleukin 2 receptor, beta | - | HPRD,BioGRID | 11133764 |
| INSR | CD220 | HHF5 | insulin receptor | Suppressors of cytokine signaling-1 associates with and inhibits the insulin receptor. | BIND | 11342531 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | - | HPRD,BioGRID | 11208867 |12228220 |
| IRS2 | - | insulin receptor substrate 2 | Affinity Capture-Western | BioGRID | 12228220 |
| ITK | EMT | LYK | MGC126257 | MGC126258 | PSCTK2 | IL2-inducible T-cell kinase | Socs1 interacts with Itk. This interaction was modeled on a demonstrated interaction between mouse Socs1 and Itk from an unspecified species. | BIND | 10022833 |
| ITK | EMT | LYK | MGC126257 | MGC126258 | PSCTK2 | IL2-inducible T-cell kinase | - | HPRD,BioGRID | 10022833 |
| JAK1 | JAK1A | JAK1B | JTK3 | Janus kinase 1 (a protein tyrosine kinase) | - | HPRD | 11133764 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | - | HPRD | 9202126 |10064597 |11971965 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | Affinity Capture-Western in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 9202126 |10064597 |10455112 |11713228 |11971965 |
| JAK3 | JAK-3 | JAK3_HUMAN | JAKL | L-JAK | LJAK | Janus kinase 3 (a protein tyrosine kinase, leukocyte) | - | HPRD | 11133764 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | - | HPRD,BioGRID | 10022833 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | Socs1 interacts with Kit. This interaction was modeled on a demonstrated interaction between mouse proteins | BIND | 10022833 |
| MPL | C-MPL | CD110 | MPLV | TPOR | myeloproliferative leukemia virus oncogene | Affinity Capture-Western | BioGRID | 10517496 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | Socs1 interacts with Nck. This interaction was modeled on a demonstrated interaction between mouse Socs1 and Nck from an unspecified species. | BIND | 10022833 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | - | HPRD,BioGRID | 10022833 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | Socs1 interacts with PDGF-R. This interaction was modeled on a demonstrated interaction between mouse Socs1 and human PDGF-R. | BIND | 10022833 |
| PIK3R2 | P85B | p85 | p85-BETA | phosphoinositide-3-kinase, regulatory subunit 2 (beta) | Socs1 interacts with p85-beta. This interaction was modeled on a demonstrated interaction between mouse Socs1 and p85-beta from an unspecified species. | BIND | 10022833 |
| TCEB1 | SIII | transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) | - | HPRD,BioGRID | 10051596 |
| TCEB2 | ELOB | SIII | transcription elongation factor B (SIII), polypeptide 2 (18kDa, elongin B) | - | HPRD | 10051596 |
| TCEB3 | FLJ38760 | FLJ42849 | SIII | TCEB3A | transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A) | Affinity Capture-Western | BioGRID | 10051596 |
| TEC | MGC126760 | MGC126762 | PSCTK4 | tec protein tyrosine kinase | - | HPRD,BioGRID | 9341160 |
| TEK | CD202B | TIE-2 | TIE2 | VMCM | VMCM1 | TEK tyrosine kinase, endothelial | Socs1 interacts with TEK. This interaction was modeled on a demonstrated interaction between mouse Socs1 and Tek from an unspecified species. | BIND | 10022833 |
| TEK | CD202B | TIE-2 | TIE2 | VMCM | VMCM1 | TEK tyrosine kinase, endothelial | - | HPRD | 10022833 |
| TRIM8 | GERP | RNF27 | tripartite motif-containing 8 | - | HPRD | 12163497 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD,BioGRID | 10022833 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | Socs1 interacts with Vav. This interaction was modeled on a demonstrated interaction between mouse Socs1 and Vav. | BIND | 10022833 |
| VAV2 | - | vav 2 guanine nucleotide exchange factor | - | HPRD,BioGRID | 10022833 |
| VAV2 | - | vav 2 guanine nucleotide exchange factor | Socs1 interacts with Vav2. This interaction was modeled on a demonstrated interaction between mouse Socs1 and human Vav2. | BIND | 10022833 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG UBIQUITIN MEDIATED PROTEOLYSIS | 138 | 98 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE II DIABETES MELLITUS | 47 | 41 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GH PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL2RB PATHWAY | 38 | 26 | All SZGR 2.0 genes in this pathway |
| SIG IL4RECEPTOR IN B LYPHOCYTES | 27 | 23 | All SZGR 2.0 genes in this pathway |
| PID IL4 2PATHWAY | 65 | 43 | All SZGR 2.0 genes in this pathway |
| PID IL12 2PATHWAY | 63 | 54 | All SZGR 2.0 genes in this pathway |
| PID IL2 1PATHWAY | 55 | 43 | All SZGR 2.0 genes in this pathway |
| PID IFNG PATHWAY | 40 | 34 | All SZGR 2.0 genes in this pathway |
| PID KIT PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY SCF KIT | 78 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME GROWTH HORMONE RECEPTOR SIGNALING | 24 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF KIT SIGNALING | 17 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF IFNG SIGNALING | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON GAMMA SIGNALING | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON ALPHA BETA SIGNALING | 64 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF IFNA SIGNALING | 24 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | 212 | 129 | All SZGR 2.0 genes in this pathway |
| FRASOR TAMOXIFEN RESPONSE UP | 51 | 36 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION UP | 211 | 136 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP | 88 | 58 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER H3K27ME3 IN NORMAL AND METHYLATED IN CANCER | 28 | 21 | All SZGR 2.0 genes in this pathway |
| OHM METHYLATED IN ADULT CANCERS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER DN | 36 | 24 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 6 | 11 | 9 | All SZGR 2.0 genes in this pathway |
| RADAEVA RESPONSE TO IFNA1 UP | 52 | 40 | All SZGR 2.0 genes in this pathway |
| BROCKE APOPTOSIS REVERSED BY IL6 | 144 | 98 | All SZGR 2.0 genes in this pathway |
| LU IL4 SIGNALING | 94 | 56 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| CHIARETTI ACUTE LYMPHOBLASTIC LEUKEMIA ZAP70 | 67 | 33 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER UP | 27 | 20 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| HOQUE METHYLATED IN CANCER | 56 | 45 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| PALOMERO GSI SENSITIVITY UP | 7 | 6 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE D7 UP | 107 | 67 | All SZGR 2.0 genes in this pathway |
| WUNDER INFLAMMATORY RESPONSE AND CHOLESTEROL UP | 58 | 38 | All SZGR 2.0 genes in this pathway |
| HOFFMANN PRE BI TO LARGE PRE BII LYMPHOCYTE UP | 36 | 24 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE UP | 105 | 75 | All SZGR 2.0 genes in this pathway |
| WORSCHECH TUMOR EVASION AND TOLEROGENICITY DN | 14 | 11 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA PR DN | 44 | 34 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| BOSCO ALLERGEN INDUCED TH2 ASSOCIATED MODULE | 151 | 86 | All SZGR 2.0 genes in this pathway |
| BOSCO TH1 CYTOTOXIC MODULE | 114 | 62 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |