Gene Page: EIF3B
Summary ?
| GeneID | 8662 |
| Symbol | EIF3B |
| Synonyms | EIF3-ETA|EIF3-P110|EIF3-P116|EIF3S9|PRT1 |
| Description | eukaryotic translation initiation factor 3 subunit B |
| Reference | MIM:603917|HGNC:HGNC:3280|Ensembl:ENSG00000106263|HPRD:06795|Vega:OTTHUMG00000022839 |
| Gene type | protein-coding |
| Map location | 7p22.3 |
| Pascal p-value | 0.004 |
| Sherlock p-value | 8.539E-4 |
| Fetal beta | -0.238 |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Guipponi_2014 | Whole Exome Sequencing analysis | 49 DNMs were identified by comparing the exome of 53 individuals with sporadic SCZ and of their non-affected parents |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| EIF3B | NM_001037283 | c.2029-1G.C | splice | NA | NA | Schizophrenia | DNM:Guipponi_2014 |
Section II. Transcriptome annotation
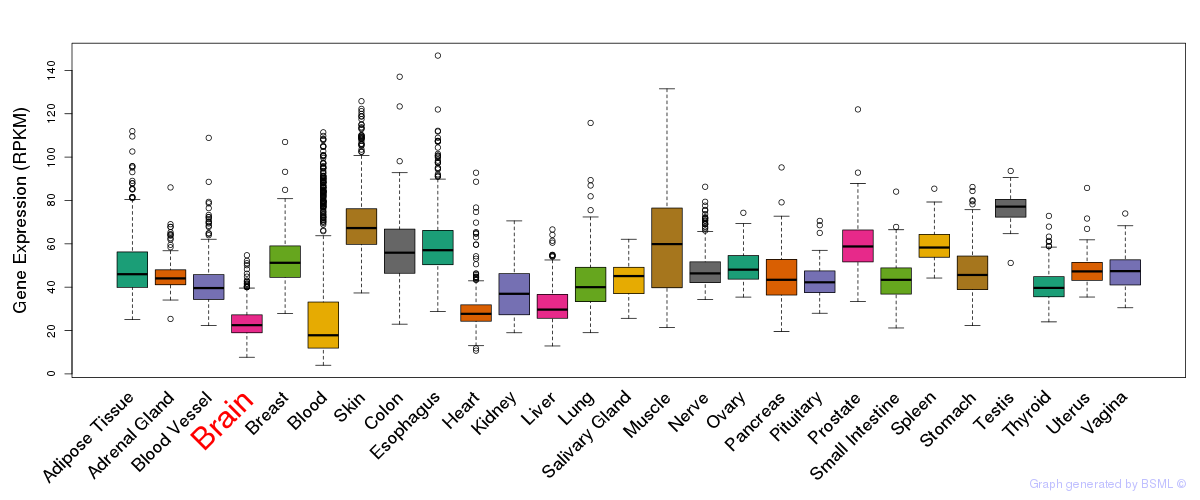
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC39A3 | 0.87 | 0.86 |
| FBXW5 | 0.86 | 0.84 |
| CDIPT | 0.86 | 0.86 |
| WDR45 | 0.85 | 0.86 |
| SLC5A2 | 0.85 | 0.82 |
| C11orf68 | 0.84 | 0.81 |
| GRINA | 0.83 | 0.82 |
| BCKDK | 0.83 | 0.81 |
| SLC41A3 | 0.83 | 0.83 |
| RABGGTA | 0.82 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.18 | -0.56 | -0.50 |
| AF347015.8 | -0.54 | -0.41 |
| MT-ATP8 | -0.54 | -0.46 |
| AF347015.21 | -0.54 | -0.42 |
| AF347015.26 | -0.52 | -0.40 |
| NSBP1 | -0.51 | -0.53 |
| MT-CO2 | -0.50 | -0.37 |
| AF347015.2 | -0.50 | -0.37 |
| EIF5B | -0.50 | -0.57 |
| AF347015.33 | -0.50 | -0.39 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| EIF3A | EIF3 | EIF3S10 | KIAA0139 | P167 | TIF32 | eIF3-p170 | eIF3-theta | p180 | p185 | eukaryotic translation initiation factor 3, subunit A | p150 interacts with hPrt1. | BIND | 11169732 |
| EIF3A | EIF3 | EIF3S10 | KIAA0139 | P167 | TIF32 | eIF3-p170 | eIF3-theta | p180 | p185 | eukaryotic translation initiation factor 3, subunit A | - | HPRD,BioGRID | 8995410 |9822659 |
| EIF3C | EIF3S8 | FLJ78287 | MGC189744 | eIF3-p110 | eukaryotic translation initiation factor 3, subunit C | Co-purification | BioGRID | 15946946 |
| EIF3I | EIF3S2 | PRO2242 | TRIP-1 | TRIP1 | eIF3-beta | eIF3-p36 | eukaryotic translation initiation factor 3, subunit I | Affinity Capture-Western | BioGRID | 16541103 |
| EIF3J | EIF3S1 | eIF3-alpha | eIF3-p35 | eukaryotic translation initiation factor 3, subunit J | Co-purification | BioGRID | 15946946 |
| EIF3K | ARG134 | EIF3-p28 | EIF3S12 | HSPC029 | M9 | MSTP001 | PLAC-24 | PLAC24 | PRO1474 | PTD001 | eukaryotic translation initiation factor 3, subunit K | - | HPRD | 14519125 |
| EIF4A1 | DDX2A | EIF-4A | EIF4A | eukaryotic translation initiation factor 4A, isoform 1 | Affinity Capture-Western | BioGRID | 16541103 |
| EIF4B | EIF-4B | PRO1843 | eukaryotic translation initiation factor 4B | Affinity Capture-Western | BioGRID | 16286006 |
| EIF4E | CBP | EIF4E1 | EIF4EL1 | EIF4F | MGC111573 | eukaryotic translation initiation factor 4E | Affinity Capture-Western | BioGRID | 16541103 |
| EIF4G1 | DKFZp686A1451 | EIF4F | EIF4G | p220 | eukaryotic translation initiation factor 4 gamma, 1 | Affinity Capture-Western | BioGRID | 16541103 |
| EIF6 | 2 | CAB | EIF3A | ITGB4BP | b | b(2)gcn | gcn | p27BBP | eukaryotic translation initiation factor 6 | Co-purification | BioGRID | 15946946 |
| FRAP1 | FLJ44809 | FRAP | FRAP2 | MTOR | RAFT1 | RAPT1 | FK506 binding protein 12-rapamycin associated protein 1 | Affinity Capture-Western | BioGRID | 16286006 |
| IL2RB | CD122 | P70-75 | interleukin 2 receptor, beta | in vivo | BioGRID | 7693677 |
| KIAA1303 | - | raptor | Affinity Capture-Western | BioGRID | 16286006 |
| RPS6 | - | ribosomal protein S6 | Affinity Capture-Western | BioGRID | 16286006 |
| RPS6KB1 | PS6K | S6K | S6K1 | STK14A | p70(S6K)-alpha | p70-S6K | p70-alpha | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | Affinity Capture-Western | BioGRID | 16286006 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRANSLATION | 222 | 75 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | 74 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | 84 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | 176 | 51 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP | 204 | 140 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| LIANG HEMATOPOIESIS STEM CELL NUMBER SMALL VS HUGE UP | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC 8HR 3 DN | 10 | 7 | All SZGR 2.0 genes in this pathway |
| WANG PROSTATE CANCER ANDROGEN INDEPENDENT | 66 | 37 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 7P22 AMPLICON | 38 | 27 | All SZGR 2.0 genes in this pathway |
| MENSSEN MYC TARGETS | 53 | 35 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION DN | 87 | 57 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| XU RESPONSE TO TRETINOIN AND NSC682994 DN | 15 | 12 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO CANTHARIDIN DN | 69 | 46 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T7 | 98 | 63 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA | 51 | 35 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |