Gene Page: B3GALNT1
Summary ?
| GeneID | 8706 |
| Symbol | B3GALNT1 |
| Synonyms | B3GALT3|GLCT3|GLOB|Gb4Cer|P|P1|beta3Gal-T3|galT3 |
| Description | beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| Reference | MIM:603094|HGNC:HGNC:918|Ensembl:ENSG00000169255|HPRD:09119|Vega:OTTHUMG00000159064 |
| Gene type | protein-coding |
| Map location | 3q25 |
| Pascal p-value | 0.036 |
| Sherlock p-value | 0.17 |
| Fetal beta | 0.13 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23252387 | 3 | 160822661 | B3GALNT1 | 3.04E-9 | -0.015 | 2.09E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs150522 | chr17 | 32701462 | B3GALNT1 | 8706 | 0.12 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
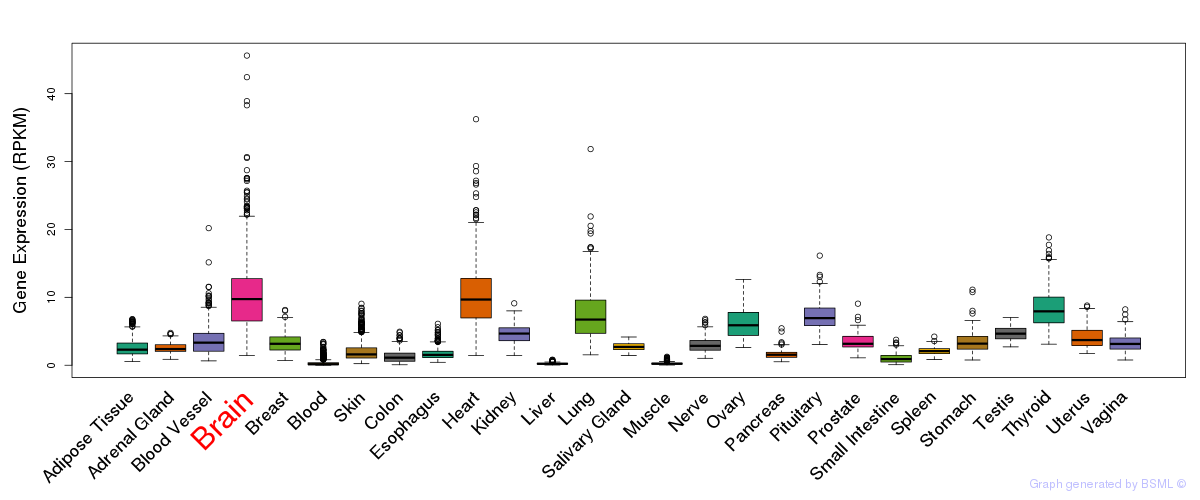
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOSPHINGOLIPID BIOSYNTHESIS GLOBO SERIES | 14 | 12 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS LOBULAR NORMAL UP | 73 | 50 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST CARCINOMA DUCTAL VS LOBULAR UP | 21 | 14 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS TOP50 DN | 45 | 26 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| XU AKT1 TARGETS 48HR | 9 | 6 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS DN | 124 | 79 | All SZGR 2.0 genes in this pathway |
| WEI MIR34A TARGETS | 148 | 97 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE UP | 283 | 177 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF DN | 103 | 64 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN | 169 | 102 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| YAO HOXA10 TARGETS VIA PROGESTERONE UP | 79 | 58 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD2 UP | 45 | 32 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART DN | 42 | 32 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS DN | 135 | 88 | All SZGR 2.0 genes in this pathway |
| TSENG ADIPOGENIC POTENTIAL DN | 46 | 28 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER DN | 101 | 65 | All SZGR 2.0 genes in this pathway |
| LIN NPAS4 TARGETS UP | 163 | 100 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN | 252 | 155 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA1 UP | 101 | 66 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |