Gene Page: RAB11A
Summary ?
| GeneID | 8766 |
| Symbol | RAB11A |
| Synonyms | YL8 |
| Description | RAB11A, member RAS oncogene family |
| Reference | MIM:605570|HGNC:HGNC:9760|Ensembl:ENSG00000103769|HPRD:05715|Vega:OTTHUMG00000133162 |
| Gene type | protein-coding |
| Map location | 15q22.31 |
| Pascal p-value | 0.442 |
| Sherlock p-value | 0.005 |
| Fetal beta | 0.472 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.8396 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23900535 | 15 | 66161693 | RAB11A | 2.14E-8 | -0.008 | 7.26E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
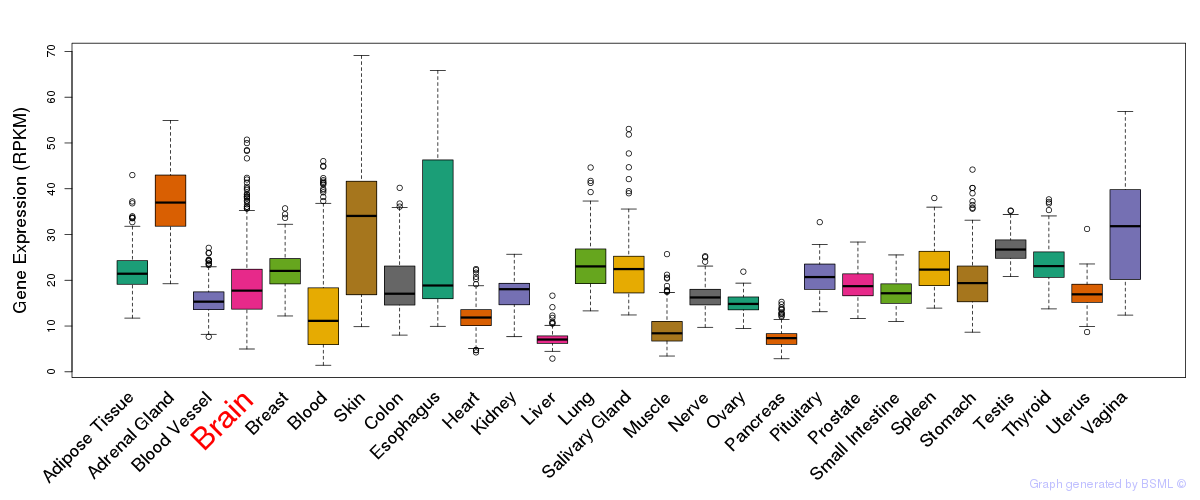
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0019905 | syntaxin binding | NAS | Synap (GO term level: 5) | 12145319 |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003924 | GTPase activity | NAS | 11163216 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0005525 | GTP binding | IEA | - | |
| GO:0005215 | transporter activity | NAS | 11163216 | |
| GO:0008134 | transcription factor binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0007264 | small GTPase mediated signal transduction | IEA | - | |
| GO:0015031 | protein transport | IEA | - | |
| GO:0048227 | plasma membrane to endosome transport | NAS | 11163216 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0055038 | recycling endosome membrane | IEA | - | |
| GO:0005802 | trans-Golgi network | IDA | 15229288 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005768 | endosome | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| GDI2 | FLJ16452 | FLJ37352 | RABGDIB | GDP dissociation inhibitor 2 | - | HPRD,BioGRID | 10512627 |
| MYO5B | KIAA1119 | myosin VB | - | HPRD,BioGRID | 11408590 |11495908 |
| RAB11FIP1 | DKFZp686E2214 | FLJ22524 | FLJ22622 | MGC78448 | NOEL1A | RCP | rab11-FIP1 | RAB11 family interacting protein 1 (class I) | Affinity Capture-MS in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 11495908 |11786538 |17353931 |
| RAB11FIP2 | KIAA0941 | Rab11-FIP2 | nRip11 | RAB11 family interacting protein 2 (class I) | - | HPRD | 15304524 |
| RAB11FIP2 | KIAA0941 | Rab11-FIP2 | nRip11 | RAB11 family interacting protein 2 (class I) | - | HPRD,BioGRID | 11495908 |11994279 |
| RAB11FIP3 | KIAA0665 | Rab11-FIP3 | RAB11 family interacting protein 3 (class II) | in vitro Two-hybrid | BioGRID | 11495908 |
| RAB11FIP3 | KIAA0665 | Rab11-FIP3 | RAB11 family interacting protein 3 (class II) | - | HPRD | 11481332 |
| RAB11FIP4 | FLJ00131 | KIAA1821 | MGC11316 | MGC126566 | RAB11-FIP4 | RAB11 family interacting protein 4 (class II) | - | HPRD,BioGRID | 12470645 |
| RAB11FIP5 | DKFZp434H018 | GAF1 | KIAA0857 | RIP11 | pp75 | RAB11 family interacting protein 5 (class I) | Rab11 interacts with the carboxy terminus, including the coiled-coil region, of Gaf-1. | BIND | 12554740 |
| RAB11FIP5 | DKFZp434H018 | GAF1 | KIAA0857 | RIP11 | pp75 | RAB11 family interacting protein 5 (class I) | Affinity Capture-Western in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 11163216 |11495908 |
| RAB3IL1 | GRAB | RAB3A interacting protein (rabin3)-like 1 | Two-hybrid | BioGRID | 16189514 |
| SEC13 | D3S1231E | SEC13L1 | SEC13R | npp-20 | SEC13 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 10747849 |
| STX4 | STX4A | p35-2 | syntaxin 4 | - | HPRD | 12435603 |
| WDR44 | DKFZp686L20145 | MGC26781 | RAB11BP | RPH11 | WD repeat domain 44 | - | HPRD,BioGRID | 10077598 |10464283 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| KEGG VASOPRESSIN REGULATED WATER REABSORPTION | 44 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RAB PATHWAY | 12 | 8 | All SZGR 2.0 genes in this pathway |
| PID NOTCH PATHWAY | 59 | 49 | All SZGR 2.0 genes in this pathway |
| PID TXA2PATHWAY | 57 | 43 | All SZGR 2.0 genes in this pathway |
| PID ARF6 DOWNSTREAM PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| PID IL8 CXCR2 PATHWAY | 34 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME AQUAPORIN MEDIATED TRANSPORT | 51 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS RED DN | 25 | 19 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 DN | 126 | 86 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 UP | 120 | 73 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| OUELLET CULTURED OVARIAN CANCER INVASIVE VS LMP UP | 69 | 40 | All SZGR 2.0 genes in this pathway |
| SEIDEN MET SIGNALING | 19 | 16 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| TOMLINS PROSTATE CANCER UP | 40 | 27 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR DN | 41 | 30 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS DN | 87 | 69 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| JIANG VHL TARGETS | 138 | 91 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION DN | 98 | 67 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D7 | 40 | 21 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| JOSEPH RESPONSE TO SODIUM BUTYRATE UP | 31 | 21 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING UP | 93 | 62 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION UP | 271 | 165 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 517 | 524 | 1A,m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 517 | 523 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-141/200a | 863 | 869 | 1A | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-142-3p | 1167 | 1173 | 1A | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-142-5p | 146 | 152 | 1A | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-181 | 226 | 232 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-21 | 363 | 370 | 1A,m8 | hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
| miR-216 | 51 | 57 | m8 | hsa-miR-216 | UAAUCUCAGCUGGCAACUGUG |
| miR-26 | 308 | 314 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-30-5p | 250 | 256 | m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-542-3p | 273 | 279 | m8 | hsa-miR-542-3p | UGUGACAGAUUGAUAACUGAAA |
| miR-543 | 227 | 233 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 1258 | 1265 | 1A,m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.