Gene Page: INPP4B
Summary ?
| GeneID | 8821 |
| Symbol | INPP4B |
| Synonyms | - |
| Description | inositol polyphosphate-4-phosphatase type II B |
| Reference | MIM:607494|HGNC:HGNC:6075|Ensembl:ENSG00000109452|HPRD:06322|Vega:OTTHUMG00000161341 |
| Gene type | protein-coding |
| Map location | 4q31.21 |
| Pascal p-value | 0.016 |
| Sherlock p-value | 0.175 |
| Fetal beta | -2.181 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03476007 | 4 | 143768727 | INPP4B | 3.794E-4 | 0.656 | 0.043 | DMG:Wockner_2014 |
| cg24759237 | 4 | 142956467 | INPP4B | 6.32E-8 | 0.014 | 1.57E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4740614 | chr9 | 15554959 | INPP4B | 8821 | 0.03 | trans | ||
| rs10962066 | chr9 | 15555037 | INPP4B | 8821 | 0.01 | trans | ||
| rs7873152 | chr9 | 15555457 | INPP4B | 8821 | 0.01 | trans | ||
| rs10962070 | chr9 | 15566016 | INPP4B | 8821 | 0.08 | trans | ||
| rs10481557 | chr9 | 15604745 | INPP4B | 8821 | 0.06 | trans | ||
| rs4741528 | chr9 | 15655977 | INPP4B | 8821 | 0.02 | trans | ||
| rs6474944 | chr9 | 15670444 | INPP4B | 8821 | 0.01 | trans | ||
| rs6474952 | chr9 | 15711233 | INPP4B | 8821 | 0.01 | trans | ||
| rs10756697 | chr9 | 15728516 | INPP4B | 8821 | 0.06 | trans | ||
| rs4740624 | chr9 | 15753410 | INPP4B | 8821 | 0.17 | trans | ||
| rs7019851 | chr9 | 15769550 | INPP4B | 8821 | 0.03 | trans | ||
| rs4146291 | chr9 | 15783265 | INPP4B | 8821 | 0.06 | trans | ||
| rs10756704 | chr9 | 15783815 | INPP4B | 8821 | 0.05 | trans | ||
| rs7032457 | chr9 | 15826729 | INPP4B | 8821 | 0.03 | trans | ||
| rs9407654 | chr9 | 15829262 | INPP4B | 8821 | 0.04 | trans | ||
| rs1891212 | chr9 | 15900770 | INPP4B | 8821 | 0.03 | trans | ||
| rs16919920 | 0 | INPP4B | 8821 | 0.17 | trans |
Section II. Transcriptome annotation
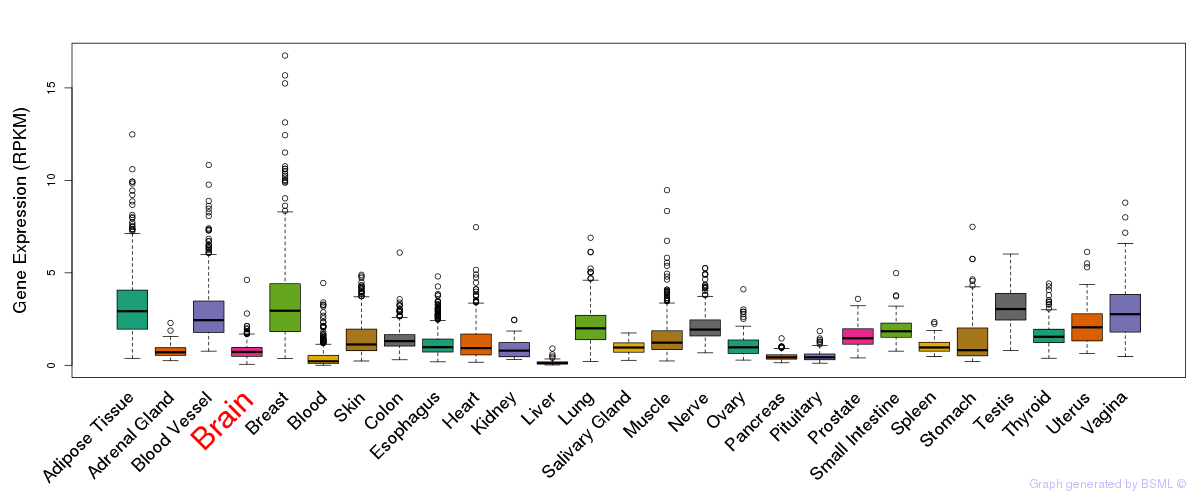
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GLT8D1 | 0.77 | 0.74 |
| C20orf12 | 0.76 | 0.79 |
| ALG13 | 0.76 | 0.77 |
| RRAGB | 0.76 | 0.78 |
| NRBP1 | 0.75 | 0.78 |
| ARMCX2 | 0.75 | 0.78 |
| PARP2 | 0.75 | 0.82 |
| FSIP1 | 0.75 | 0.75 |
| VPS33B | 0.75 | 0.77 |
| TPST1 | 0.74 | 0.70 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.67 | -0.65 |
| AF347015.8 | -0.67 | -0.66 |
| AF347015.33 | -0.67 | -0.64 |
| AF347015.27 | -0.66 | -0.66 |
| AF347015.2 | -0.66 | -0.65 |
| AF347015.31 | -0.65 | -0.64 |
| AF347015.15 | -0.65 | -0.64 |
| MT-CYB | -0.65 | -0.63 |
| AF347015.26 | -0.65 | -0.66 |
| AF347015.21 | -0.62 | -0.63 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG INOSITOL PHOSPHATE METABOLISM | 54 | 42 | All SZGR 2.0 genes in this pathway |
| KEGG PHOSPHATIDYLINOSITOL SIGNALING SYSTEM | 76 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | 12 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPID METABOLISM | 198 | 112 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | 31 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME PI METABOLISM | 48 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN | 182 | 111 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS UP | 94 | 57 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 UP | 112 | 72 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 6HR UP | 85 | 54 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 3D UP | 182 | 110 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D UP | 194 | 122 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| STREICHER LSM1 TARGETS UP | 44 | 34 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP | 207 | 128 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| YANAGIHARA ESX1 TARGETS | 30 | 19 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 UP | 36 | 22 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| LIN SILENCED BY TUMOR MICROENVIRONMENT | 108 | 73 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| NELSON RESPONSE TO ANDROGEN UP | 86 | 61 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS TARGETS UP | 84 | 51 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR UP | 78 | 56 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| GEORGANTAS HSC MARKERS | 71 | 47 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN THYMUS UP | 196 | 137 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE UP | 97 | 61 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| LEE NAIVE T LYMPHOCYTE | 19 | 10 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN | 134 | 83 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |