Gene Page: DLGAP1
Summary ?
| GeneID | 9229 |
| Symbol | DLGAP1 |
| Synonyms | DAP-1|DAP-1-ALPHA|DAP-1-BETA|DAP1|DLGAP1A|DLGAP1B|GKAP|SAPAP1 |
| Description | discs large homolog associated protein 1 |
| Reference | MIM:605445|HGNC:HGNC:2905|Ensembl:ENSG00000170579|HPRD:09260|Vega:OTTHUMG00000131537 |
| Gene type | protein-coding |
| Map location | 18p11.31 |
| Pascal p-value | 0.46 |
| Fetal beta | -1.616 |
| Support | PROTEIN CLUSTERING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_Synaptosome G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanNRC CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
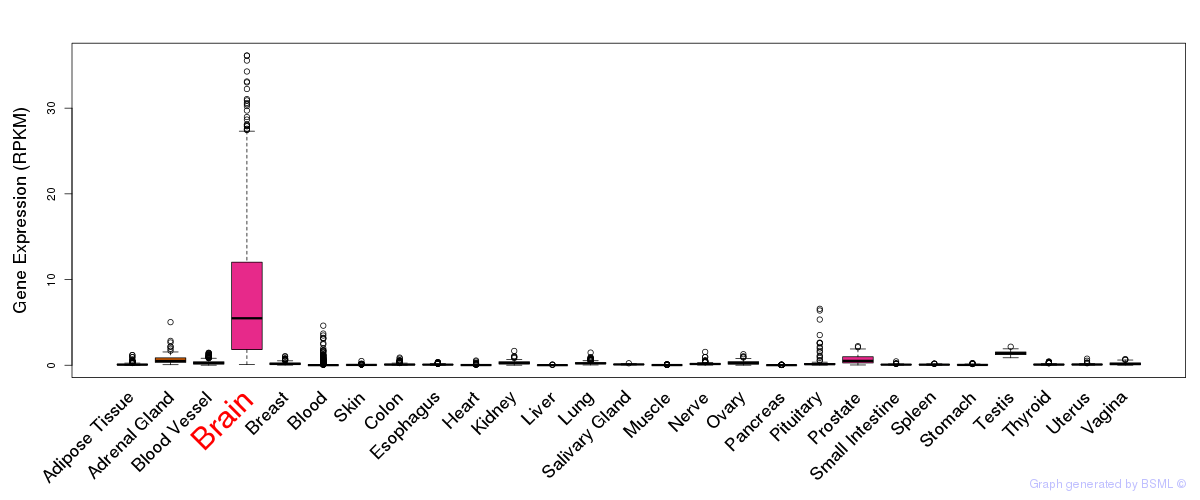
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SPTLC1 | 0.75 | 0.73 |
| MRPL19 | 0.75 | 0.71 |
| PAIP2 | 0.74 | 0.72 |
| SCP2 | 0.73 | 0.71 |
| RABL3 | 0.72 | 0.70 |
| CEPT1 | 0.72 | 0.69 |
| PCNP | 0.72 | 0.67 |
| TBC1D15 | 0.71 | 0.66 |
| ADK | 0.71 | 0.72 |
| C6orf211 | 0.71 | 0.70 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AL022328.1 | -0.40 | -0.37 |
| AC135724.1 | -0.39 | -0.41 |
| IL32 | -0.39 | -0.22 |
| AF347015.18 | -0.38 | -0.15 |
| CXCL1 | -0.36 | -0.30 |
| AF347015.21 | -0.35 | -0.06 |
| FAM159B | -0.34 | -0.17 |
| RP9P | -0.34 | -0.29 |
| C10orf108 | -0.34 | -0.30 |
| AC010300.1 | -0.33 | -0.24 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | TAS | 9024696 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | NAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 9286858 |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APC | BTPS2 | DP2 | DP2.5 | DP3 | GS | adenomatous polyposis coli | - | HPRD,BioGRID | 9286858 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Affinity Capture-Western | BioGRID | 9286858 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | - | HPRD,BioGRID | 9286858 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | hDLG interacts with DAP-1alpha. | BIND | 9286858 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | DLGAP1 (GKAP) interacts with DLG1 (SAP97). | BIND | 15729360 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | hDLG interacts with DAP-1beta. | BIND | 9286858 |
| DLG2 | DKFZp781D1854 | DKFZp781E0954 | FLJ37266 | MGC131811 | PSD-93 | discs, large homolog 2, chapsyn-110 (Drosophila) | DAP-1 interacts with Chapsyn-110. | BIND | 11122378 |
| DLG2 | DKFZp781D1854 | DKFZp781E0954 | FLJ37266 | MGC131811 | PSD-93 | discs, large homolog 2, chapsyn-110 (Drosophila) | Affinity Capture-Western | BioGRID | 10844022 |
| DLG3 | KIAA1232 | MRX | MRX90 | NE-Dlg | NEDLG | SAP102 | discs, large homolog 3 (neuroendocrine-dlg, Drosophila) | Two-hybrid | BioGRID | 9115257 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | Affinity Capture-Western Co-fractionation Far Western Reconstituted Complex Two-hybrid | BioGRID | 9024696 |9115257 |9286858 |10527873 |10844022 |11060025 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD | 9024696 |9286858 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | PSD-95 interacts with DAP-1alpha. | BIND | 9286858 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 10844022 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | DAP-1 interacts with DLC-1. | BIND | 11122378 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | DLC8 interacts with GKAP. This interaction was modeled on a demonstrated interaction between DLC8 from an unspecified species and human GKAP. | BIND | 11148209 |
| DYNLL2 | DNCL1B | Dlc2 | MGC17810 | dynein, light chain, LC8-type 2 | DAP-1 interacts with DLC-2. | BIND | 11122378 |
| DYNLL2 | DNCL1B | Dlc2 | MGC17810 | dynein, light chain, LC8-type 2 | - | HPRD,BioGRID | 10844022 |
| GRIN2A | NMDAR2A | NR2A | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | - | HPRD,BioGRID | 9286858 |
| KCNA4 | HBK4 | HK1 | HPCN2 | HUKII | KCNA4L | KCNA8 | KV1.4 | PCN2 | potassium voltage-gated channel, shaker-related subfamily, member 4 | - | HPRD | 9024696 |
| MYO5A | GS1 | MYH12 | MYO5 | MYR12 | myosin VA (heavy chain 12, myoxin) | Reconstituted Complex | BioGRID | 10844022 |
| SHANK1 | SPANK-1 | SSTRIP | synamon | SH3 and multiple ankyrin repeat domains 1 | Co-crystal Structure | BioGRID | 12954649 |
| SHANK2 | CORTBP1 | CTTNBP1 | ProSAP1 | SHANK | SPANK-3 | SH3 and multiple ankyrin repeat domains 2 | - | HPRD,BioGRID | 10527873 |
| SHANK3 | KIAA1650 | PROSAP2 | PSAP2 | SPANK-2 | SH3 and multiple ankyrin repeat domains 3 | Two-hybrid | BioGRID | 10527873 |
| SORBS2 | ARGBP2 | FLJ93447 | KIAA0777 | PRO0618 | sorbin and SH3 domain containing 2 | - | HPRD,BioGRID | 10521485 |
| TANC1 | KIAA1728 | ROLSB | TANC | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | - | HPRD | 15673434 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ASTON MAJOR DEPRESSIVE DISORDER UP | 49 | 36 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND UP | 77 | 52 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP | 222 | 139 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER WITH H3K27ME3 | 196 | 93 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME2 | 17 | 11 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3 UNMETHYLATED | 228 | 119 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-146 | 335 | 341 | m8 | hsa-miR-146a | UGAGAACUGAAUUCCAUGGGUU |
| hsa-miR-146bbrain | UGAGAACUGAAUUCCAUAGGCU | ||||
| miR-423 | 210 | 216 | m8 | hsa-miR-423 | AGCUCGGUCUGAGGCCCCUCAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.