Gene Page: CYTH2
Summary ?
| GeneID | 9266 |
| Symbol | CYTH2 |
| Synonyms | ARNO|CTS18|CTS18.1|PSCD2|PSCD2L|SEC7L|Sec7p-L|Sec7p-like |
| Description | cytohesin 2 |
| Reference | MIM:602488|HGNC:HGNC:9502|Ensembl:ENSG00000105443|HPRD:03925|Vega:OTTHUMG00000150245 |
| Gene type | protein-coding |
| Map location | 19q13.33 |
| Pascal p-value | 0.454 |
| Sherlock p-value | 0.848 |
| Fetal beta | 0.866 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0089 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24083067 | 19 | 48972665 | CYTH2 | 7.94E-5 | -0.54 | 0.025 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
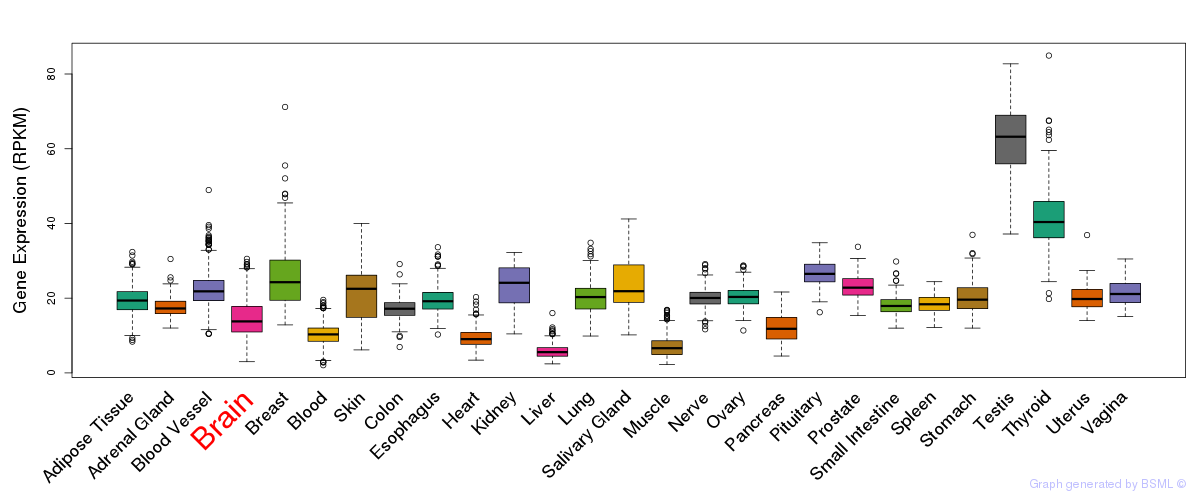
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NPY1R | 0.74 | 0.59 |
| C1orf62 | 0.72 | 0.59 |
| LMO4 | 0.72 | 0.55 |
| TAL2 | 0.68 | 0.57 |
| NPNT | 0.66 | 0.52 |
| RASGEF1C | 0.65 | 0.66 |
| RPRM | 0.65 | 0.63 |
| PTX3 | 0.64 | 0.57 |
| STK32B | 0.63 | 0.54 |
| PPP1R14C | 0.63 | 0.61 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| REEP6 | -0.43 | -0.42 |
| TSC22D4 | -0.39 | -0.42 |
| AIFM3 | -0.39 | -0.42 |
| AF347015.27 | -0.39 | -0.49 |
| HLA-F | -0.39 | -0.39 |
| RHBDL3 | -0.39 | -0.47 |
| HEPN1 | -0.38 | -0.41 |
| RENBP | -0.38 | -0.43 |
| CLU | -0.38 | -0.39 |
| AF347015.33 | -0.38 | -0.49 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005086 | ARF guanyl-nucleotide exchange factor activity | IEA | - | |
| GO:0005086 | ARF guanyl-nucleotide exchange factor activity | TAS | 9417041 | |
| GO:0005515 | protein binding | IPI | 14654833 |16415858 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006897 | endocytosis | TAS | 9417041 | |
| GO:0030036 | actin cytoskeleton organization | TAS | 9417041 | |
| GO:0032012 | regulation of ARF protein signal transduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005624 | membrane fraction | TAS | 9417041 | |
| GO:0005886 | plasma membrane | TAS | 9417041 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA ARAP PATHWAY | 18 | 13 | All SZGR 2.0 genes in this pathway |
| PID ARF6 PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI PATHWAY | 49 | 40 | All SZGR 2.0 genes in this pathway |
| PID ARF 3PATHWAY | 19 | 13 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| ALCALA APOPTOSIS | 88 | 60 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| PETRETTO BLOOD PRESSURE DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR UP | 37 | 31 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C2 | 25 | 18 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA EARLY RESPONSE TO TGFB1 | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE | 206 | 117 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 1HR DN | 10 | 8 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 | 182 | 102 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS DN | 187 | 109 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| ZWANG DOWN BY 2ND EGF PULSE | 293 | 119 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-28 | 114 | 120 | m8 | hsa-miR-28brain | AAGGAGCUCACAGUCUAUUGAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.