Gene Page: NRXN2
Summary ?
| GeneID | 9379 |
| Symbol | NRXN2 |
| Synonyms | - |
| Description | neurexin 2 |
| Reference | MIM:600566|HGNC:HGNC:8009|Ensembl:ENSG00000110076|HPRD:11859|Vega:OTTHUMG00000045214 |
| Gene type | protein-coding |
| Map location | 11q13 |
| Pascal p-value | 0.062 |
| Fetal beta | -0.493 |
| DMG | 2 (# studies) |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 4 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 4 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NRXN2 | chr11 | 64435116 | C | T | NM_015080 NM_138732 | . . | silent silent | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg27243166 | 11 | 64479496 | NRXN2 | 1.26E-6 | 0.48 | 0.007 | DMG:Wockner_2014 |
| cg03035661 | 11 | 64409932 | NRXN2 | 2.27E-5 | 0.894 | 0.017 | DMG:Wockner_2014 |
| cg26388730 | 11 | 64405764 | NRXN2 | 8.96E-5 | 0.348 | 0.027 | DMG:Wockner_2014 |
| cg21996761 | 11 | 64428434 | NRXN2 | 2.96E-8 | 0.017 | 9.1E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
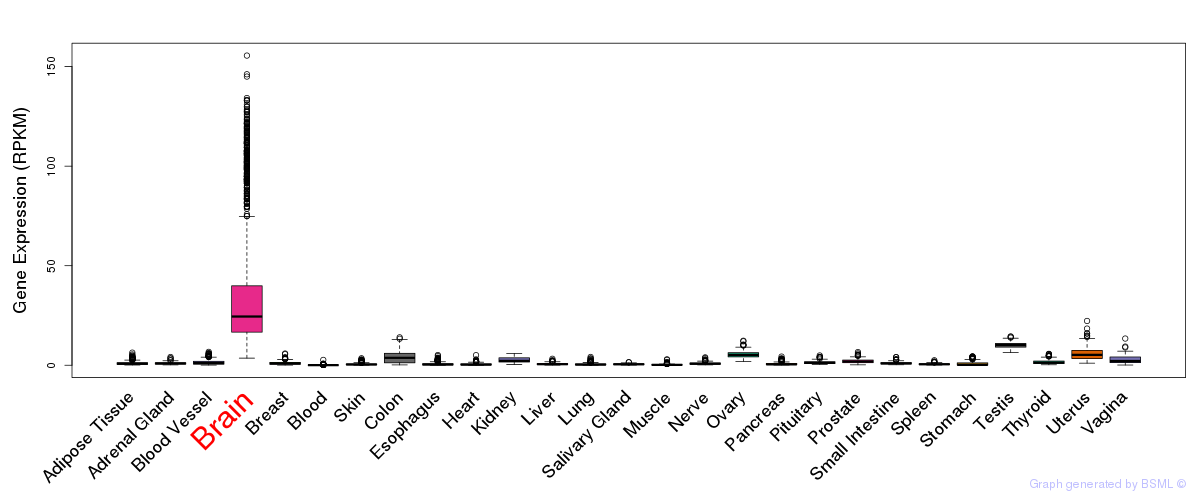
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAD23B | 0.94 | 0.93 |
| CS | 0.94 | 0.94 |
| EHBP1 | 0.93 | 0.94 |
| PDXDC1 | 0.93 | 0.93 |
| IARS | 0.93 | 0.93 |
| C4orf41 | 0.93 | 0.93 |
| AP1G1 | 0.93 | 0.94 |
| RABGEF1 | 0.93 | 0.92 |
| WDR47 | 0.93 | 0.95 |
| CUL1 | 0.93 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.83 | -0.85 |
| MT-CO2 | -0.83 | -0.85 |
| AF347015.8 | -0.81 | -0.84 |
| AF347015.33 | -0.79 | -0.82 |
| AF347015.21 | -0.79 | -0.84 |
| AF347015.27 | -0.79 | -0.83 |
| FXYD1 | -0.79 | -0.83 |
| MT-CYB | -0.79 | -0.81 |
| HIGD1B | -0.79 | -0.83 |
| AF347015.2 | -0.78 | -0.83 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN | 182 | 111 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO HD MTX DN | 24 | 18 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR DN | 63 | 41 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME2 | 17 | 11 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING UP | 93 | 62 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |