Gene Page: TGFBRAP1
Summary ?
| GeneID | 9392 |
| Symbol | TGFBRAP1 |
| Synonyms | TRAP-1|TRAP1|VPS3 |
| Description | transforming growth factor beta receptor associated protein 1 |
| Reference | MIM:606237|HGNC:HGNC:16836|Ensembl:ENSG00000135966|HPRD:06948|Vega:OTTHUMG00000130809 |
| Gene type | protein-coding |
| Map location | 2q12.1 |
| Pascal p-value | 0.027 |
| Fetal beta | -0.091 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0004 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00755 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14459716 | 2 | 105889560 | TGFBRAP1 | 3.926E-4 | 0.458 | 0.043 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7595584 | chr2 | 175652187 | TGFBRAP1 | 9392 | 0.04 | trans | ||
| rs17029291 | chr3 | 32402138 | TGFBRAP1 | 9392 | 0.02 | trans | ||
| rs830202 | chr3 | 165654796 | TGFBRAP1 | 9392 | 0.01 | trans | ||
| rs17160578 | chr7 | 85672108 | TGFBRAP1 | 9392 | 0.09 | trans | ||
| rs722970 | chr7 | 85803264 | TGFBRAP1 | 9392 | 0.04 | trans | ||
| rs17324099 | chr7 | 85856288 | TGFBRAP1 | 9392 | 0.08 | trans | ||
| rs12569493 | chr10 | 74886042 | TGFBRAP1 | 9392 | 0.1 | trans | ||
| rs16930362 | chr10 | 74890320 | TGFBRAP1 | 9392 | 0.04 | trans | ||
| rs7092445 | chr10 | 74921991 | TGFBRAP1 | 9392 | 0.13 | trans | ||
| rs16915029 | chr10 | 75001994 | TGFBRAP1 | 9392 | 0.1 | trans | ||
| rs12571454 | chr10 | 75208954 | TGFBRAP1 | 9392 | 0.15 | trans | ||
| rs16958358 | chr17 | 55502410 | TGFBRAP1 | 9392 | 0.02 | trans | ||
| rs1761450 | chr19 | 54818034 | TGFBRAP1 | 9392 | 0.04 | trans | ||
| rs6032295 | chr20 | 44178138 | TGFBRAP1 | 9392 | 0.15 | trans | ||
| rs6098023 | chr20 | 53113740 | TGFBRAP1 | 9392 | 0.13 | trans | ||
| rs17264923 | chrX | 69499164 | TGFBRAP1 | 9392 | 0.05 | trans |
Section II. Transcriptome annotation
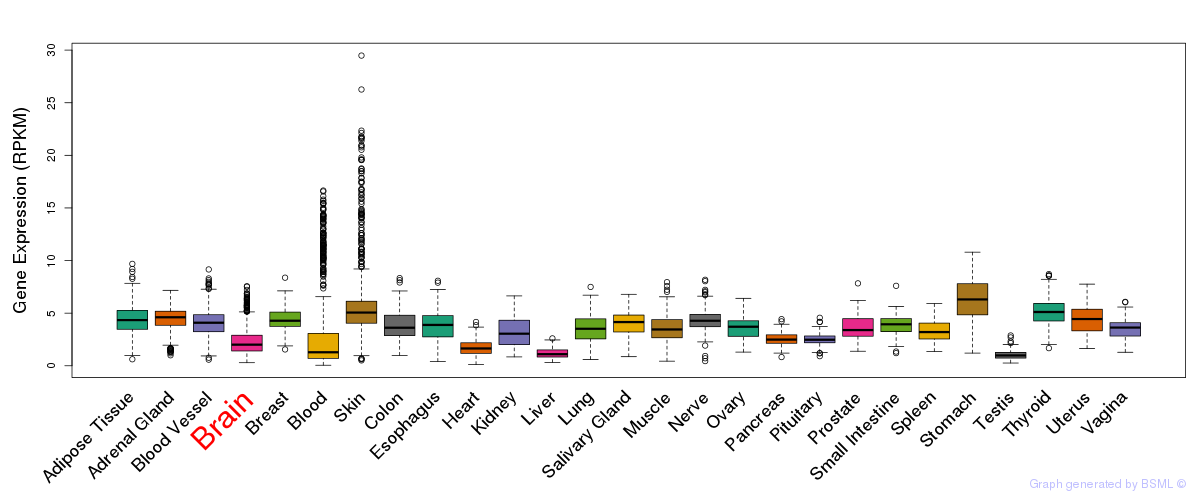
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NOP56 | 0.88 | 0.88 |
| DDX56 | 0.87 | 0.85 |
| TRAPPC3 | 0.87 | 0.85 |
| WIBG | 0.87 | 0.87 |
| EXOSC7 | 0.86 | 0.86 |
| PSMD13 | 0.86 | 0.87 |
| NGDN | 0.86 | 0.88 |
| KAT5 | 0.86 | 0.87 |
| C1orf174 | 0.86 | 0.85 |
| BCS1L | 0.85 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.78 | -0.74 |
| AF347015.8 | -0.77 | -0.76 |
| MT-CYB | -0.77 | -0.76 |
| AF347015.2 | -0.77 | -0.77 |
| AF347015.31 | -0.76 | -0.75 |
| AF347015.27 | -0.76 | -0.77 |
| AF347015.15 | -0.75 | -0.76 |
| AF347015.26 | -0.75 | -0.77 |
| AF347015.33 | -0.75 | -0.74 |
| AF347015.9 | -0.70 | -0.73 |
Section III. Gene Ontology annotation
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACVR1B | ACTRIB | ACVRLK4 | ALK4 | SKR2 | activin A receptor, type IB | - | HPRD | 11278302 |
| ACVR2B | ACTRIIB | ActR-IIB | MGC116908 | activin A receptor, type IIB | - | HPRD | 11278302 |
| ACVRL1 | ACVRLK1 | ALK-1 | ALK1 | HHT | HHT2 | ORW2 | SKR3 | TSR-I | activin A receptor type II-like 1 | - | HPRD | 11278302 |
| ALOX5 | 5-LO | 5-LOX | 5LPG | LOG5 | MGC163204 | arachidonate 5-lipoxygenase | - | HPRD,BioGRID | 10051563 |
| ALOX5 | 5-LO | 5-LOX | 5LPG | LOG5 | MGC163204 | arachidonate 5-lipoxygenase | 5LO interacts with TRAP-1. | BIND | 10051563 |
| BMPR1A | 10q23del | ACVRLK3 | ALK3 | CD292 | bone morphogenetic protein receptor, type IA | - | HPRD | 11278302 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD | 11278302 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | - | HPRD,BioGRID | 11278302 |
| TGFB1 | CED | DPD1 | TGFB | TGFbeta | transforming growth factor, beta 1 | Affinity Capture-Western | BioGRID | 11278302 |
| TGFB2 | MGC116892 | TGF-beta2 | transforming growth factor, beta 2 | Affinity Capture-Western | BioGRID | 11278302 |
| TGFBR1 | AAT5 | ACVRLK4 | ALK-5 | ALK5 | LDS1A | LDS2A | SKR4 | TGFR-1 | transforming growth factor, beta receptor 1 | - | HPRD | 11278302 |
| TGFBR2 | AAT3 | FAA3 | LDS1B | LDS2B | MFS2 | RIIC | TAAD2 | TGFR-2 | TGFbeta-RII | transforming growth factor, beta receptor II (70/80kDa) | - | HPRD | 11278302 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID SMAD2 3PATHWAY | 17 | 13 | All SZGR 2.0 genes in this pathway |
| PID TGFBR PATHWAY | 55 | 38 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR UP | 111 | 68 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING VIA ATM NOT VIA NFKB DN | 38 | 23 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |