Gene Page: ARHGAP32
Summary ?
| GeneID | 9743 |
| Symbol | ARHGAP32 |
| Synonyms | GC-GAP|GRIT|PX-RICS|RICS|p200RhoGAP|p250GAP |
| Description | Rho GTPase activating protein 32 |
| Reference | MIM:608541|HGNC:HGNC:17399|Ensembl:ENSG00000134909|HPRD:12257|Vega:OTTHUMG00000165774 |
| Gene type | protein-coding |
| Map location | 11q24.3 |
| Pascal p-value | 0.015 |
| Sherlock p-value | 0.341 |
| Fetal beta | -1.094 |
| eGene | Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10894158 | chr11 | 129736156 | ARHGAP32 | 9743 | 0.09 | cis |
Section II. Transcriptome annotation
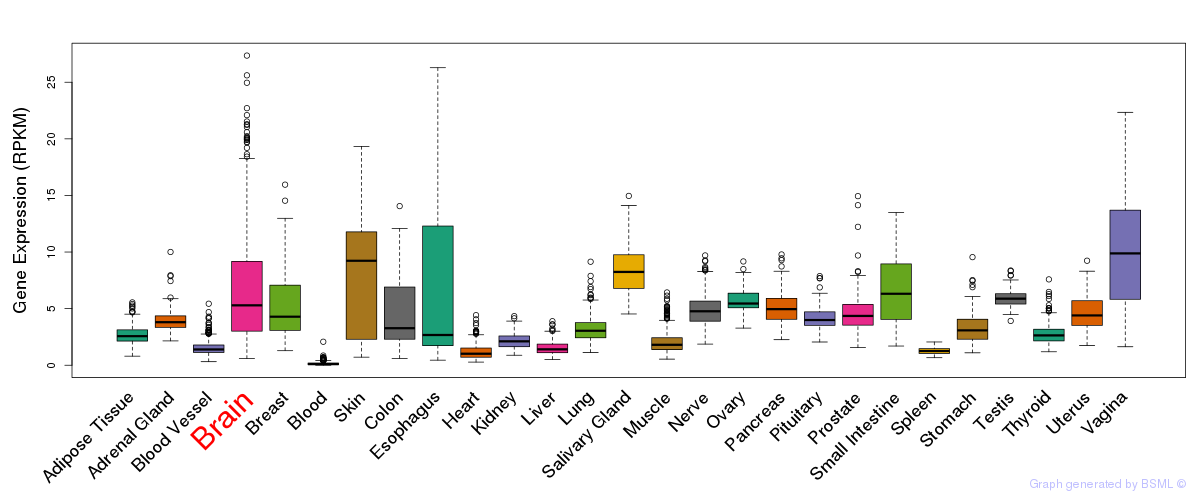
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ANKRD17 | 0.93 | 0.94 |
| ADAR | 0.93 | 0.94 |
| DYRK1A | 0.93 | 0.94 |
| ANAPC1 | 0.93 | 0.94 |
| NOLC1 | 0.93 | 0.93 |
| KIAA0090 | 0.93 | 0.93 |
| SLC12A6 | 0.92 | 0.95 |
| FTO | 0.92 | 0.93 |
| HERC1 | 0.92 | 0.93 |
| ARHGEF7 | 0.92 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.77 | -0.79 |
| MT-CO2 | -0.77 | -0.81 |
| AF347015.31 | -0.76 | -0.79 |
| HIGD1B | -0.76 | -0.81 |
| AF347015.21 | -0.74 | -0.83 |
| IFI27 | -0.73 | -0.77 |
| C1orf54 | -0.72 | -0.85 |
| AF347015.33 | -0.72 | -0.74 |
| AC021016.1 | -0.72 | -0.74 |
| MT-CYB | -0.71 | -0.75 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005096 | GTPase activator activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0035091 | phosphoinositide binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007165 | signal transduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042995 | cell projection | IEA | axon (GO term level: 4) | - |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0000139 | Golgi membrane | IEA | - | |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005789 | endoplasmic reticulum membrane | IEA | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005768 | endosome | IEA | - | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0010008 | endosome membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | Biochemical Activity | BioGRID | 12446789 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | - | HPRD,BioGRID | 12446789 |12819203 |
| CAMK2N2 | CAM-KIIN | CAMKIIN | calcium/calmodulin-dependent protein kinase II inhibitor 2 | - | HPRD | 12531901 |
| CDC42 | CDC42Hs | G25K | cell division cycle 42 (GTP binding protein, 25kDa) | Biochemical Activity | BioGRID | 12446789 |12819203 |12857875 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | - | HPRD | 12531901 |
| CDH2 | CD325 | CDHN | CDw325 | NCAD | cadherin 2, type 1, N-cadherin (neuronal) | - | HPRD | 12531901 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | Crk interacts with GC-GAP. This interaction was modelled on a demonstrated interaction between Crk from an unspecified species and human GC-GAP. | BIND | 12819203 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | Affinity Capture-Western Reconstituted Complex | BioGRID | 12454018 |12819203 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | - | HPRD | 12446789 |12454018 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | - | HPRD,BioGRID | 12446789 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD | 12531901 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD | 12531901 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 12788081 |12857875 |
| GAB1 | - | GRB2-associated binding protein 1 | Gab1 interacts with GC-GAP. | BIND | 12819203 |
| GAB1 | - | GRB2-associated binding protein 1 | - | HPRD,BioGRID | 12819203 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | Gab2 interacts with GC-GAP. | BIND | 12819203 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | - | HPRD,BioGRID | 12819203 |
| GRAP | MGC64880 | GRB2-related adaptor protein | Grap interacts with GC-GAP. This interaction was modelled on a demonstrated interaction between Grap from an unspecified species and human GC-GAP. | BIND | 12819203 |
| GRIN2A | NMDAR2A | NR2A | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | - | HPRD | 12531901 |
| GRIN2B | MGC142178 | MGC142180 | NMDAR2B | NR2B | hNR3 | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | - | HPRD,BioGRID | 12531901 |12857875 |
| MEGF10 | DKFZp781K1852 | FLJ41574 | KIAA1780 | multiple EGF-like-domains 10 | - | HPRD | 12421765 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | Nck interacts with GC-GAP. This interaction was modelled on a demonstrated interaction between Nck from an unspecified species and human GC-GAP. | BIND | 12819203 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | - | HPRD,BioGRID | 12819203 |
| NF2 | ACN | BANF | SCH | neurofibromin 2 (merlin) | Affinity Capture-Western | BioGRID | 12446789 |
| NTRK1 | DKFZp781I14186 | MTC | TRK | TRK1 | TRKA | p140-TrkA | neurotrophic tyrosine kinase, receptor, type 1 | - | HPRD,BioGRID | 12446789 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD,BioGRID | 12454018 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD,BioGRID | 12454018 |
| RAC1 | MGC111543 | MIG5 | TC-25 | p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | Biochemical Activity | BioGRID | 12454018 |12819203 |
| RHOA | ARH12 | ARHA | RHO12 | RHOH12 | ras homolog gene family, member A | Biochemical Activity | BioGRID | 12446789 |12454018 |12857875 |
| SFN | YWHAS | stratifin | Affinity Capture-MS | BioGRID | 15778465 |
| SHC2 | SCK | SHCB | SLI | SHC (Src homology 2 domain containing) transforming protein 2 | Affinity Capture-Western | BioGRID | 12446789 |
| SHC3 | N-Shc | NSHC | RAI | SHCC | SHC (Src homology 2 domain containing) transforming protein 3 | - | HPRD,BioGRID | 12446789 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 12454018 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Src interacts with GC-GAP. This interaction was modelled on a demonstrated interaction between Src from an unspecified species and human GC-GAP. | BIND | 12819203 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED MODERATELY VS POORLY UP | 121 | 71 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE DN | 204 | 114 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN VINCRISTINE RESISTANCE ALL UP | 27 | 16 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-132/212 | 44 | 50 | m8 | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-137 | 9 | 15 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-338 | 32 | 38 | 1A | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| miR-485-5p | 25 | 31 | m8 | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.