Gene Page: TELO2
Summary ?
| GeneID | 9894 |
| Symbol | TELO2 |
| Synonyms | CLK2|TEL2 |
| Description | telomere maintenance 2 |
| Reference | MIM:611140|HGNC:HGNC:29099|Ensembl:ENSG00000100726|HPRD:10016|Vega:OTTHUMG00000044471 |
| Gene type | protein-coding |
| Map location | 16p13.3 |
| Pascal p-value | 0.972 |
| Sherlock p-value | 0.376 |
| Fetal beta | -0.035 |
| DMG | 1 (# studies) |
| eGene | Cortex Frontal Cortex BA9 Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08699794 | 16 | 1557513 | TELO2 | 4.268E-4 | 0.41 | 0.045 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | TELO2 | 9894 | 0.02 | trans | ||
| rs16955618 | chr15 | 29937543 | TELO2 | 9894 | 0.01 | trans |
Section II. Transcriptome annotation
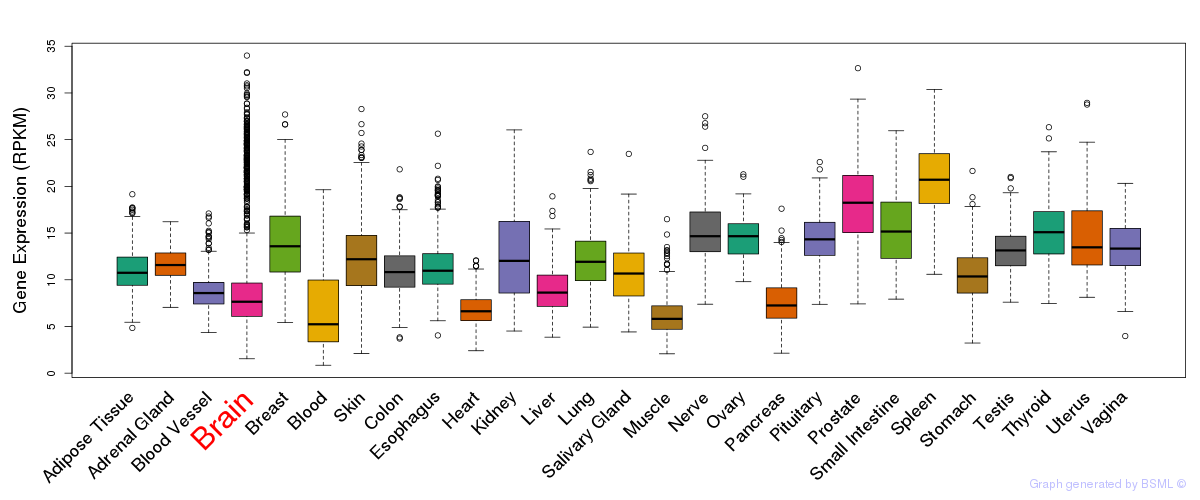
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TNK2 | 0.79 | 0.78 |
| SLC6A8 | 0.79 | 0.77 |
| PYCR2 | 0.79 | 0.80 |
| ALAD | 0.78 | 0.75 |
| SLC48A1 | 0.78 | 0.79 |
| BDH1 | 0.78 | 0.81 |
| AGFG2 | 0.77 | 0.79 |
| PDK2 | 0.77 | 0.70 |
| PNPO | 0.76 | 0.75 |
| DHRS11 | 0.76 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| YBX1 | -0.48 | -0.49 |
| CCDC112 | -0.48 | -0.38 |
| RBMX2 | -0.48 | -0.50 |
| KCNRG | -0.47 | -0.36 |
| AC016138.1 | -0.47 | -0.45 |
| IDH1 | -0.47 | -0.39 |
| PHF14 | -0.47 | -0.48 |
| C11orf57 | -0.47 | -0.48 |
| TTC27 | -0.47 | -0.38 |
| SQLE | -0.47 | -0.39 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| GARCIA TARGETS OF FLI1 AND DAX1 DN | 176 | 104 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 16P13 AMPLICON | 120 | 49 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| LIN APC TARGETS | 77 | 55 | All SZGR 2.0 genes in this pathway |
| GALE APL WITH FLT3 MUTATED UP | 56 | 35 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP | 202 | 115 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |