Gene Page: MED13
Summary ?
| GeneID | 9969 |
| Symbol | MED13 |
| Synonyms | ARC250|DRIP250|HSPC221|THRAP1|TRAP240 |
| Description | mediator complex subunit 13 |
| Reference | MIM:603808|HGNC:HGNC:22474|Ensembl:ENSG00000108510|HPRD:07229|Vega:OTTHUMG00000179251 |
| Gene type | protein-coding |
| Map location | 17q23.2 |
| Pascal p-value | 0.079 |
| Fetal beta | 1.795 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| Expression | Meta-analysis of gene expression | P value: 1.38 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20376447 | 17 | 60143446 | MED13 | 1.18E-9 | -0.029 | 1.28E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12620093 | chr2 | 69393712 | MED13 | 9969 | 0.1 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
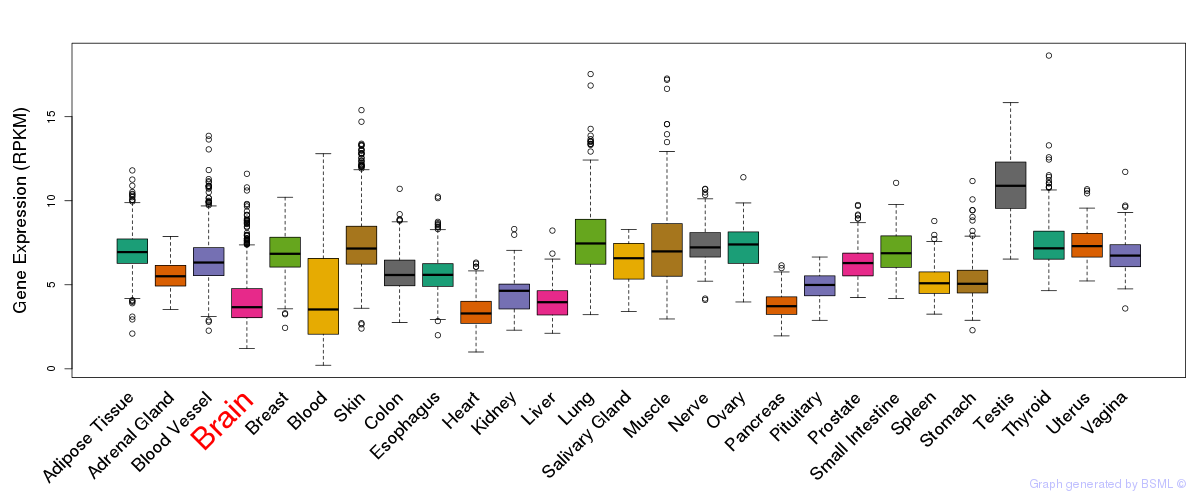
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DMXL1 | 0.93 | 0.95 |
| NF1 | 0.93 | 0.95 |
| GARNL1 | 0.92 | 0.92 |
| ATG2B | 0.92 | 0.93 |
| CXorf39 | 0.92 | 0.92 |
| DMXL2 | 0.91 | 0.92 |
| AC010896.1 | 0.91 | 0.95 |
| APPL1 | 0.91 | 0.92 |
| SEL1L | 0.91 | 0.94 |
| C9orf5 | 0.91 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.59 | -0.58 |
| AF347015.21 | -0.58 | -0.56 |
| C1orf61 | -0.57 | -0.70 |
| C1orf54 | -0.57 | -0.62 |
| HIGD1B | -0.57 | -0.59 |
| MT-CO2 | -0.55 | -0.56 |
| CSAG1 | -0.55 | -0.55 |
| VAMP5 | -0.55 | -0.53 |
| AC098691.2 | -0.55 | -0.59 |
| EIF4EBP3 | -0.54 | -0.57 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IDA | 12218053 | |
| GO:0016455 | RNA polymerase II transcription mediator activity | IDA | 10198638 | |
| GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity | NAS | 10235266 | |
| GO:0042809 | vitamin D receptor binding | NAS | 10235266 | |
| GO:0046966 | thyroid hormone receptor binding | IDA | 10198638 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | IDA | 12218053 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0030521 | androgen receptor signaling pathway | IDA | 12218053 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IDA | 12037571 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000119 | mediator complex | IDA | 10198638 | |
| GO:0005634 | nucleus | IDA | 10235267 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS D UP | 38 | 23 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER UP | 181 | 108 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER CLUSTER 5 | 18 | 11 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 3084 | 3090 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-128 | 760 | 766 | m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU | ||||
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-129-5p | 3818 | 3824 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-134 | 263 | 270 | 1A,m8 | hsa-miR-134brain | UGUGACUGGUUGACCAGAGGG |
| miR-135 | 2742 | 2748 | m8 | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA | ||||
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA | ||||
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA | ||||
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-136 | 329 | 335 | 1A | hsa-miR-136 | ACUCCAUUUGUUUUGAUGAUGGA |
| miR-137 | 538 | 544 | m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-140 | 2893 | 2899 | m8 | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-142-5p | 3623 | 3629 | m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-144 | 3083 | 3090 | 1A,m8 | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-145 | 3525 | 3531 | m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU | ||||
| miR-146 | 1450 | 1456 | 1A | hsa-miR-146a | UGAGAACUGAAUUCCAUGGGUU |
| hsa-miR-146bbrain | UGAGAACUGAAUUCCAUAGGCU | ||||
| miR-15/16/195/424/497 | 1595 | 1601 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-153 | 2737 | 2743 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-17-5p/20/93.mr/106/519.d | 3622 | 3628 | 1A | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-18 | 2796 | 2802 | 1A | hsa-miR-18a | UAAGGUGCAUCUAGUGCAGAUA |
| hsa-miR-18b | UAAGGUGCAUCUAGUGCAGUUA | ||||
| miR-183 | 3740 | 3746 | 1A | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
| miR-19 | 1531 | 1538 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-194 | 1017 | 1023 | 1A | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-196 | 1664 | 1670 | 1A | hsa-miR-196a | UAGGUAGUUUCAUGUUGUUGG |
| hsa-miR-196b | UAGGUAGUUUCCUGUUGUUGG | ||||
| miR-200bc/429 | 2659 | 2665 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC | ||||
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC | ||||
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-205 | 3628 | 3634 | m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-208 | 565 | 572 | 1A,m8 | hsa-miR-208 | AUAAGACGAGCAAAAAGCUUGU |
| miR-217 | 582 | 588 | 1A | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-24 | 288 | 294 | 1A | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-26 | 1641 | 1647 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC | ||||
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-27 | 2761 | 2768 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-299-5p | 2892 | 2898 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-323 | 3034 | 3040 | m8 | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-324-3p | 3657 | 3663 | 1A | hsa-miR-324-3p | CCACUGCCCCAGGUGCUGCUGG |
| miR-34b | 521 | 527 | m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| miR-369-3p | 603 | 609 | m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU | ||||
| miR-374 | 222 | 229 | 1A,m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-375 | 3717 | 3723 | 1A | hsa-miR-375 | UUUGUUCGUUCGGCUCGCGUGA |
| miR-379 | 1662 | 1668 | m8 | hsa-miR-379brain | UGGUAGACUAUGGAACGUA |
| miR-381 | 390 | 396 | m8 | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-410 | 894 | 900 | m8 | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| hsa-miR-410 | AAUAUAACACAGAUGGCCUGU | ||||
| hsa-miR-410 | AAUAUAACACAGAUGGCCUGU | ||||
| miR-433-3p | 3850 | 3856 | 1A | hsa-miR-433brain | AUCAUGAUGGGCUCCUCGGUGU |
| miR-448 | 2737 | 2743 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-485-3p | 2776 | 2783 | 1A,m8 | hsa-miR-485-3p | GUCAUACACGGCUCUCCUCUCU |
| miR-493-5p | 742 | 748 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU | ||||
| hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU | ||||
| miR-499 | 566 | 572 | 1A | hsa-miR-499 | UUAAGACUUGCAGUGAUGUUUAA |
| miR-543 | 1015 | 1021 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.