Gene Page: MPP6
Summary ?
| GeneID | 51678 |
| Symbol | MPP6 |
| Synonyms | PALS2|VAM-1|VAM1|p55T |
| Description | membrane protein, palmitoylated 6 |
| Reference | MIM:606959|HGNC:HGNC:18167|Ensembl:ENSG00000105926|HPRD:09509|Vega:OTTHUMG00000023507 |
| Gene type | protein-coding |
| Map location | 7p15 |
| Pascal p-value | 0.002 |
| Fetal beta | 0.465 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
| Support | PROTEIN CLUSTERING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.05 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10678266 | 7 | 24614348 | MPP6 | 2.275E-4 | 0.615 | 0.036 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2813827 | chr7 | 24252970 | MPP6 | 51678 | 0.18 | cis | ||
| rs2642265 | chr7 | 24253061 | MPP6 | 51678 | 0.15 | cis |
Section II. Transcriptome annotation
General gene expression (GTEx)
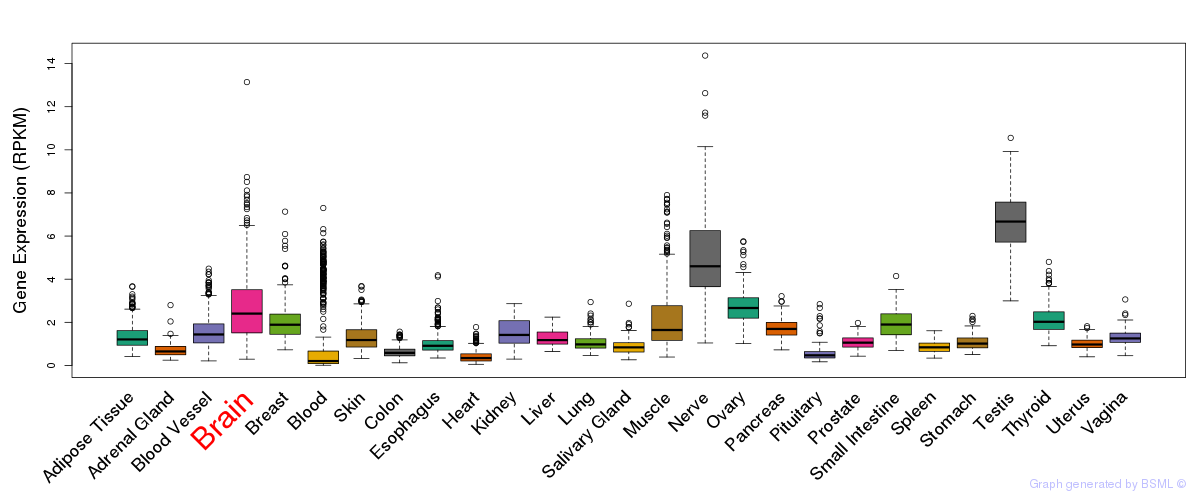
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 11311936 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006461 | protein complex assembly | NAS | 11311936 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AATF | CHE-1 | CHE1 | DED | apoptosis antagonizing transcription factor | Two-hybrid | BioGRID | 15231747 |
| ARHGAP18 | FLJ25728 | MGC126757 | MGC138145 | MacGAP | bA307O14.2 | Rho GTPase activating protein 18 | Two-hybrid | BioGRID | 15231747 |
| C15orf23 | FLJ14502 | HSD11 | MGC141728 | MGC141729 | TRAF4AF1 | chromosome 15 open reading frame 23 | Two-hybrid | BioGRID | 15231747 |
| DNM2 | CMTDI1 | CMTDIB | DI-CMTB | DYN2 | DYNII | dynamin 2 | - | HPRD,BioGRID | 15231747 |
| DYNLRB1 | BITH | BLP | DNCL2A | DNLC2A | ROBLD1 | dynein, light chain, roadblock-type 1 | - | HPRD,BioGRID | 15231747 |
| EIF3G | EIF3-P42 | EIF3S4 | eIF3-delta | eIF3-p44 | eukaryotic translation initiation factor 3, subunit G | - | HPRD,BioGRID | 15231747 |
| EXOSC10 | PM-Scl | PM/Scl-100 | PMSCL | PMSCL2 | RRP6 | Rrp6p | p2 | p3 | p4 | exosome component 10 | Two-hybrid | BioGRID | 15231747 |
| FTL | MGC71996 | ferritin, light polypeptide | - | HPRD,BioGRID | 15231747 |
| KCNJ12 | FLJ14167 | IRK2 | KCNJN1 | Kir2.2 | Kir2.2v | hIRK | hIRK1 | hkir2.2x | kcnj12x | potassium inwardly-rectifying channel, subfamily J, member 12 | Affinity Capture-MS | BioGRID | 15024025 |
| LIN7A | LIN-7A | LIN7 | MALS-1 | MGC148143 | TIP-33 | VELI1 | lin-7 homolog A (C. elegans) | - | HPRD,BioGRID | 10753959 |11311936 |
| RPS20 | FLJ27451 | MGC102930 | ribosomal protein S20 | - | HPRD,BioGRID | 15231747 |
| SKIV2L2 | Dob1 | KIAA0052 | MGC142069 | Mtr4 | fSAP118 | superkiller viralicidic activity 2-like 2 (S. cerevisiae) | Two-hybrid | BioGRID | 15231747 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | - | HPRD,BioGRID | 15231747 |
| SNX9 | MST155 | MSTP155 | SDP1 | SH3PX1 | SH3PXD3A | WISP | sorting nexin 9 | Two-hybrid | BioGRID | 15231747 |
| THOP1 | EP24.15 | MEPD_HUMAN | MP78 | TOP | thimet oligopeptidase 1 | - | HPRD,BioGRID | 15231747 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DOANE BREAST CANCER ESR1 DN | 48 | 28 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 LASER DN | 50 | 38 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| SU TESTIS | 76 | 53 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER CLUSTER 3 | 16 | 12 | All SZGR 2.0 genes in this pathway |