Gene Page: NOG
Summary ?
| GeneID | 9241 |
| Symbol | NOG |
| Synonyms | SYM1|SYNS1|SYNS1A |
| Description | noggin |
| Reference | MIM:602991|HGNC:HGNC:7866|Ensembl:ENSG00000183691|HPRD:04291|Vega:OTTHUMG00000151770 |
| Gene type | protein-coding |
| Map location | 17q22 |
| Pascal p-value | 0.143 |
| Fetal beta | 0.182 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:PheWAS | Phenome-wide association studies (PheWAS) | 157 SNPs associated with schizophrenia | 0 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 CV:PheWAS
CV:PheWAS
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|
Section II. Transcriptome annotation
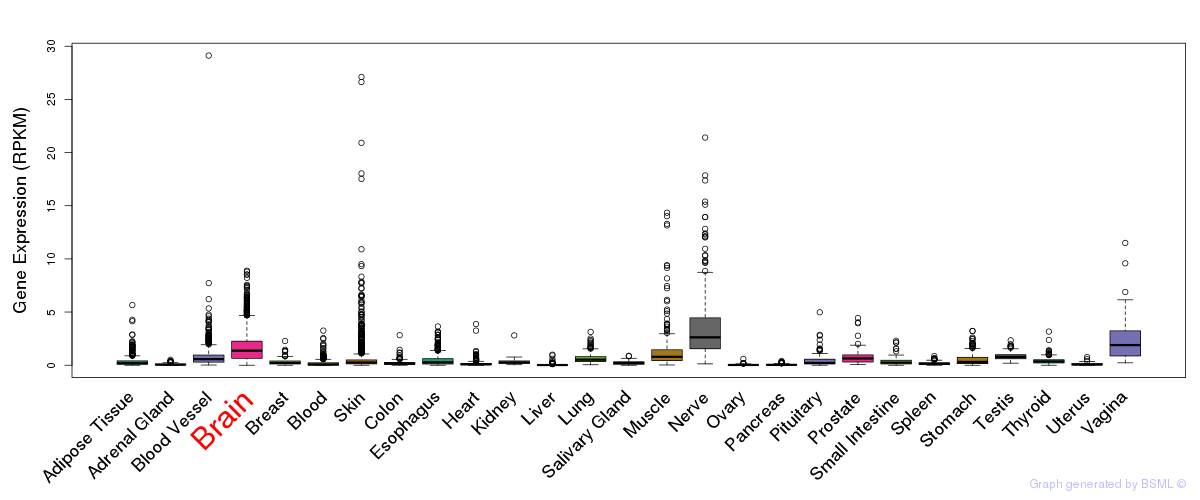
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC6A20 | 0.81 | 0.56 |
| SLC22A2 | 0.75 | 0.63 |
| SLC13A4 | 0.73 | 0.78 |
| SLC5A5 | 0.69 | 0.77 |
| OGN | 0.66 | 0.69 |
| DSP | 0.62 | 0.56 |
| SERPIND1 | 0.61 | 0.69 |
| GJB2 | 0.60 | 0.60 |
| SLC22A6 | 0.59 | 0.69 |
| OMD | 0.59 | 0.54 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LRRC37A | -0.18 | -0.36 |
| CHMP4A | -0.17 | -0.30 |
| NBR2 | -0.17 | -0.20 |
| C11orf51 | -0.14 | -0.23 |
| ACP6 | -0.14 | -0.06 |
| USF2 | -0.13 | -0.31 |
| ZRSR2 | -0.13 | -0.36 |
| HES4 | -0.13 | -0.23 |
| SLC25A28 | -0.13 | -0.17 |
| C14orf80 | -0.13 | -0.15 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0019955 | cytokine binding | IPI | 8752214 | |
| GO:0042803 | protein homodimerization activity | IDA | 11562478 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | IEA | Brain (GO term level: 7) | - |
| GO:0048712 | negative regulation of astrocyte differentiation | ISS | astrocyte, Glial (GO term level: 12) | 10780858 |
| GO:0001657 | ureteric bud development | IEA | - | |
| GO:0001649 | osteoblast differentiation | ISS | 10780858 | |
| GO:0001837 | epithelial to mesenchymal transition | ISS | 10780858 | |
| GO:0009953 | dorsal/ventral pattern formation | IDA | 7666191 | |
| GO:0007605 | sensory perception of sound | IEA | - | |
| GO:0007389 | pattern specification process | IEA | - | |
| GO:0042060 | wound healing | ISS | 10780858 | |
| GO:0035019 | somatic stem cell maintenance | IMP | 17889703 | |
| GO:0042733 | embryonic digit morphogenesis | IMP | 10080184 | |
| GO:0030514 | negative regulation of BMP signaling pathway | IEA | - | |
| GO:0042474 | middle ear morphogenesis | IMP | 10080184 | |
| GO:0051216 | cartilage development | IEA | - | |
| GO:0045596 | negative regulation of cell differentiation | IEA | - | |
| GO:0048570 | notochord morphogenesis | IEA | - | |
| GO:0048706 | embryonic skeletal system development | IMP | 10080184 | |
| GO:0048646 | anatomical structure formation | IEA | - | |
| GO:0060044 | negative regulation of cardiac muscle cell proliferation | ISS | 10780858 | |
| GO:0060173 | limb development | IMP | 10080184 | |
| GO:0060272 | embryonic skeletal joint morphogenesis | IMP | 16151340 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | EXP | 17029022 | |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | TAS | 10080184 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ALK PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| PID BMP PATHWAY | 42 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY BMP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS DN | 123 | 78 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D UP | 157 | 91 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS UP | 127 | 79 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT UP | 197 | 110 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| WONG IFNA2 RESISTANCE DN | 34 | 20 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| LEE NAIVE T LYMPHOCYTE | 19 | 10 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K4ME3 AND H3K27ME3 | 210 | 148 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-148/152 | 172 | 178 | m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-181 | 192 | 198 | 1A | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-200bc/429 | 247 | 254 | 1A,m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-329 | 204 | 210 | 1A | hsa-miR-329brain | AACACACCUGGUUAACCUCUUU |
| miR-369-3p | 249 | 256 | 1A,m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 250 | 256 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.