Gene Page: NOTCH4
Summary ?
| GeneID | 4855 |
| Symbol | NOTCH4 |
| Synonyms | INT3 |
| Description | notch 4 |
| Reference | MIM:164951|HGNC:HGNC:7884|Ensembl:ENSG00000204301|HPRD:01290|Vega:OTTHUMG00000031044 |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 1E-12 |
| Sherlock p-value | 0.15 |
| Fetal beta | -0.546 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26950898 | 6 | 32164380 | NOTCH4 | 9.49E-8 | -0.007 | 2.14E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7132043 | chr12 | 80968399 | NOTCH4 | 4855 | 0.19 | trans |
Section II. Transcriptome annotation
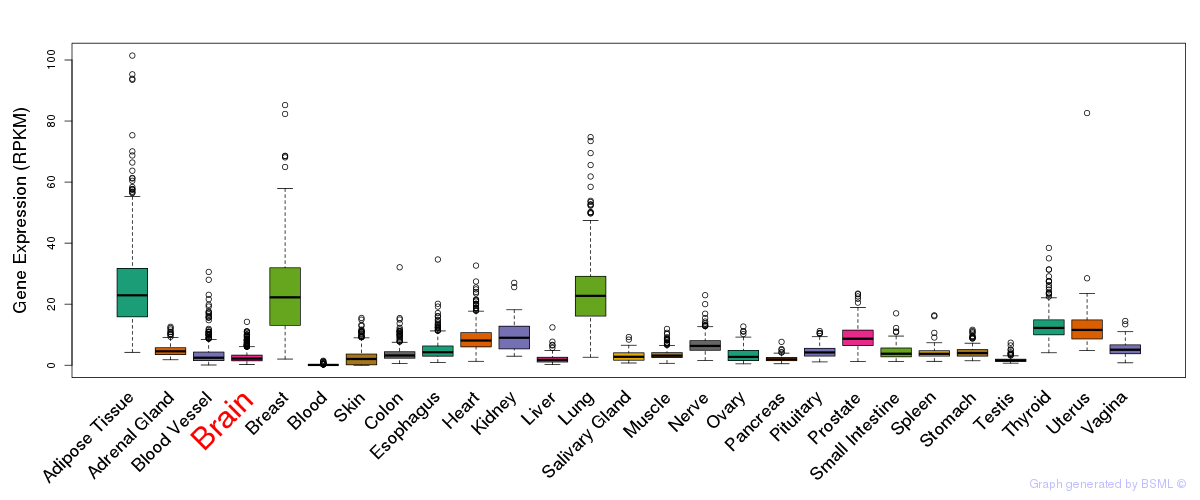
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NPY5R | 0.76 | 0.86 |
| PROM1 | 0.75 | 0.63 |
| NPNT | 0.75 | 0.62 |
| GRAP2 | 0.74 | 0.59 |
| MARCH1 | 0.74 | 0.82 |
| ARHGAP15 | 0.73 | 0.60 |
| LMO4 | 0.73 | 0.84 |
| AC079061.1 | 0.72 | 0.84 |
| GRM7 | 0.72 | 0.79 |
| RBM24 | 0.72 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| REEP6 | -0.50 | -0.47 |
| UBTD1 | -0.48 | -0.59 |
| HSD17B14 | -0.47 | -0.63 |
| RHBDL3 | -0.46 | -0.59 |
| AIFM3 | -0.46 | -0.57 |
| TSC22D4 | -0.45 | -0.58 |
| S100B | -0.45 | -0.59 |
| BDH2 | -0.44 | -0.59 |
| ACSF2 | -0.44 | -0.64 |
| RENBP | -0.44 | -0.52 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | TAS | 11823422 | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005509 | calcium ion binding | TAS | 11823422 | |
| GO:0005515 | protein binding | IPI | 7671825 | |
| GO:0046982 | protein heterodimerization activity | TAS | 8681805 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001569 | patterning of blood vessels | ISS | - | |
| GO:0001709 | cell fate determination | TAS | 11823422 | |
| GO:0001763 | morphogenesis of a branching structure | ISS | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0007219 | Notch signaling pathway | IEA | - | |
| GO:0009790 | embryonic development | ISS | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0030879 | mammary gland development | IDA | 9576833 | |
| GO:0050793 | regulation of developmental process | IEA | - | |
| GO:0030097 | hemopoiesis | TAS | 11532344 | |
| GO:0045602 | negative regulation of endothelial cell differentiation | ISS | - | |
| GO:0045893 | positive regulation of transcription, DNA-dependent | TAS | 11823422 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 8681805 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0009986 | cell surface | IDA | 11823422 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 11823422 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG DORSO VENTRAL AXIS FORMATION | 25 | 21 | All SZGR 2.0 genes in this pathway |
| KEGG NOTCH SIGNALING PATHWAY | 47 | 35 | All SZGR 2.0 genes in this pathway |
| PID NOTCH PATHWAY | 59 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | 29 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH4 | 12 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME PRE NOTCH EXPRESSION AND PROCESSING | 44 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME PRE NOTCH PROCESSING IN GOLGI | 16 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | 13 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | 12 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| LIU CDX2 TARGETS UP | 36 | 22 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL UP | 121 | 72 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN | 196 | 131 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| THEODOROU MAMMARY TUMORIGENESIS | 31 | 24 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 7 | 27 | 22 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED FREQUENTLY | 12 | 10 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| GOLDRATH IMMUNE MEMORY | 65 | 42 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR DN | 21 | 13 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY UP | 86 | 48 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 24HR | 20 | 13 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 LCP WITH H3K4ME3 | 162 | 80 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K4ME3 | 174 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES LCP WITH H3K4ME3 | 142 | 80 | All SZGR 2.0 genes in this pathway |
| FUKUSHIMA TNFSF11 TARGETS | 16 | 14 | All SZGR 2.0 genes in this pathway |
| KOHOUTEK CCNT2 TARGETS | 58 | 35 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |