Gene Page: CDK5
Summary ?
| GeneID | 1020 |
| Symbol | CDK5 |
| Synonyms | LIS7|PSSALRE |
| Description | cyclin-dependent kinase 5 |
| Reference | MIM:123831|HGNC:HGNC:1774|HPRD:00449| |
| Gene type | protein-coding |
| Map location | 7q36 |
| Pascal p-value | 7.015E-4 |
| Sherlock p-value | 0.188 |
| Fetal beta | -0.927 |
| DMG | 1 (# studies) |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 14 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.152 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11368578 | 7 | 150754936 | CDK5 | 4.21E-10 | -0.016 | 7.8E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
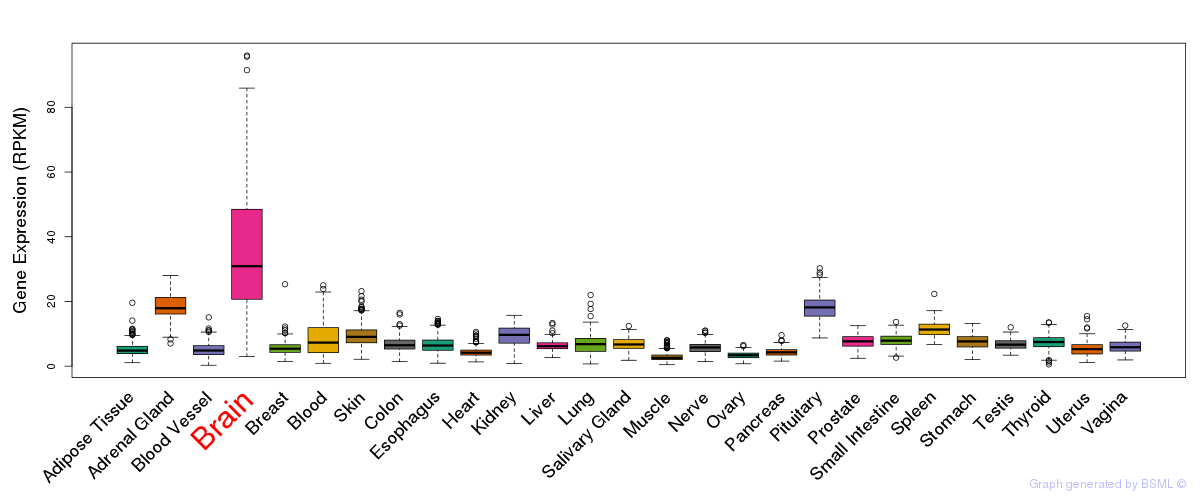
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C1R | 0.66 | 0.76 |
| PTH1R | 0.64 | 0.68 |
| PLCD3 | 0.62 | 0.72 |
| ANKRD9 | 0.59 | 0.64 |
| SDSL | 0.58 | 0.63 |
| GNPTG | 0.57 | 0.66 |
| SERTAD1 | 0.57 | 0.66 |
| OCEL1 | 0.57 | 0.60 |
| ARID5A | 0.56 | 0.66 |
| LDHD | 0.56 | 0.61 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF551 | -0.48 | -0.51 |
| AC004017.1 | -0.48 | -0.52 |
| ZNF435 | -0.48 | -0.55 |
| ZNF124 | -0.47 | -0.54 |
| MLLT3 | -0.47 | -0.49 |
| ZNF302 | -0.47 | -0.54 |
| KIAA1949 | -0.47 | -0.36 |
| ZNF766 | -0.47 | -0.53 |
| ZNF34 | -0.47 | -0.51 |
| PPAPDC1B | -0.47 | -0.50 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004693 | cyclin-dependent protein kinase activity | TAS | neuron (GO term level: 8) | 8090221 |
| GO:0050321 | tau-protein kinase activity | ISS | Brain (GO term level: 7) | - |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005176 | ErbB-2 class receptor binding | ISS | - | |
| GO:0005515 | protein binding | IPI | 12223541 | |
| GO:0005515 | protein binding | ISS | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0030549 | acetylcholine receptor activator activity | ISS | - | |
| GO:0043125 | ErbB-3 class receptor binding | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007409 | axonogenesis | IEA | neuron, axon, neurite (GO term level: 12) | - |
| GO:0008045 | motor axon guidance | IEA | neuron, axon (GO term level: 14) | - |
| GO:0030182 | neuron differentiation | ISS | neuron (GO term level: 8) | - |
| GO:0060078 | regulation of postsynaptic membrane potential | IEA | neuron, Synap, Neurotransmitter (GO term level: 9) | - |
| GO:0014044 | Schwann cell development | IEA | neuron, axon, Glial (GO term level: 10) | - |
| GO:0031175 | neurite development | ISS | neuron, axon, neurite, dendrite (GO term level: 10) | - |
| GO:0043525 | positive regulation of neuron apoptosis | ISS | neuron (GO term level: 9) | - |
| GO:0007160 | cell-matrix adhesion | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0008283 | cell proliferation | TAS | 8090221 | |
| GO:0009790 | embryonic development | ISS | - | |
| GO:0051301 | cell division | IEA | - | |
| GO:0030334 | regulation of cell migration | IEA | - | |
| GO:0043113 | receptor clustering | IEA | - | |
| GO:0045860 | positive regulation of protein kinase activity | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043025 | cell soma | ISS | axon, dendrite (GO term level: 4) | - |
| GO:0030175 | filopodium | IEA | axon (GO term level: 5) | - |
| GO:0031594 | neuromuscular junction | ISS | neuron, axon, Synap, Neurotransmitter (GO term level: 3) | - |
| GO:0030426 | growth cone | ISS | axon, dendrite (GO term level: 5) | - |
| GO:0030425 | dendrite | ISS | neuron, axon, dendrite (GO term level: 6) | - |
| GO:0030424 | axon | ISS | neuron, axon, Neurotransmitter (GO term level: 6) | - |
| GO:0005829 | cytosol | IEA | - | |
| GO:0005634 | nucleus | ISS | - | |
| GO:0005737 | cytoplasm | ISS | - | |
| GO:0016020 | membrane | ISS | - | |
| GO:0030027 | lamellipodium | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Reconstituted Complex | BioGRID | 9244350 |
| CABLES1 | CABLES | FLJ35924 | HsT2563 | IK3-1 | Cdk5 and Abl enzyme substrate 1 | - | HPRD | 10896159 |11733001 |11955625 |
| CABLES2 | C20orf150 | dJ908M14.2 | ik3-2 | Cdk5 and Abl enzyme substrate 2 | - | HPRD,BioGRID | 11955625 |
| CCND2 | KIAK0002 | MGC102758 | cyclin D2 | - | HPRD,BioGRID | 9422379 |
| CCNG1 | CCNG | cyclin G1 | - | HPRD | 9013862 |
| CDK5 | PSSALRE | cyclin-dependent kinase 5 | - | HPRD,BioGRID | 11583627 |
| CDK5R1 | CDK5P35 | CDK5R | MGC33831 | NCK5A | p23 | p25 | p35 | p35nck5a | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | p35 interacts with Cdk5. This interaction was modeled on a demonstrated interaction between bovine p35 and human Cdk5. | BIND | 9038181 |
| CDK5R1 | CDK5P35 | CDK5R | MGC33831 | NCK5A | p23 | p25 | p35 | p35nck5a | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | - | HPRD,BioGRID | 11583627 |
| CDK5R1 | CDK5P35 | CDK5R | MGC33831 | NCK5A | p23 | p25 | p35 | p35nck5a | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | Cdk5 interacts with p35. | BIND | 8090221 |
| CDK5R2 | NCK5AI | P39 | p39nck5ai | cyclin-dependent kinase 5, regulatory subunit 2 (p39) | - | HPRD,BioGRID | 10683146 |
| CDK5RAP1 | C20orf34 | C42 | CDK5RAP1.3 | CDK5RAP1.4 | CGI-05 | HSPC167 | CDK5 regulatory subunit associated protein 1 | - | HPRD,BioGRID | 11882646 |
| CDK5RAP2 | C48 | Cep215 | DKFZp686B1070 | DKFZp686D1070 | KIAA1633 | MCPH3 | CDK5 regulatory subunit associated protein 2 | Reconstituted Complex | BioGRID | 10915792 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD | 11168528 |
| FSD1 | GLFND | MGC3213 | MIR1 | fibronectin type III and SPRY domain containing 1 | - | HPRD | 12154070 |
| GAK | FLJ16629 | FLJ40395 | MGC99654 | cyclin G associated kinase | - | HPRD,BioGRID | 9013862 |
| LMTK2 | AATYK2 | BREK | KIAA1079 | KPI-2 | KPI2 | LMR2 | cprk | lemur tyrosine kinase 2 | Reconstituted Complex Two-hybrid | BioGRID | 12832520 |
| NDEL1 | DKFZp451M0318 | EOPA | MITAP1 | NUDEL | nudE nuclear distribution gene E homolog (A. nidulans)-like 1 | Biochemical Activity Co-localization | BioGRID | 11163260 |
| NES | FLJ21841 | Nbla00170 | nestin | - | HPRD,BioGRID | 12832492 |
| PAK1 | MGC130000 | MGC130001 | PAKalpha | p21 protein (Cdc42/Rac)-activated kinase 1 | Affinity Capture-Western Biochemical Activity | BioGRID | 11604394 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | - | HPRD | 7949095 |
| PCTK1 | FLJ16665 | PCTAIRE | PCTAIRE1 | PCTGAIRE | PCTAIRE protein kinase 1 | Biochemical Activity | BioGRID | 12084709 |
| STX1A | HPC-1 | STX1 | p35-1 | syntaxin 1A (brain) | - | HPRD,BioGRID | 9478941 |
| STXBP1 | EIEE4 | MUNC18-1 | UNC18 | hUNC18 | rbSec1 | syntaxin binding protein 1 | - | HPRD,BioGRID | 12963086 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BIOPEPTIDES PATHWAY | 45 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P35ALZHEIMERS PATHWAY | 11 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CDK5 PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RAC1 PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CK1 PATHWAY | 17 | 17 | All SZGR 2.0 genes in this pathway |
| PID REELIN PATHWAY | 29 | 29 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID LIS1 PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| PID EPHA FWDPATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID MAPK TRK PATHWAY | 34 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME OPIOID SIGNALLING | 78 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME DARPP 32 EVENTS | 25 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME CRMPS IN SEMA3A SIGNALING | 14 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMAPHORIN INTERACTIONS | 68 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | 132 | 101 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR DN | 64 | 39 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR DN | 75 | 50 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR DN | 21 | 13 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C4 | 20 | 14 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-142-5p | 158 | 164 | m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.