Gene Page: SF3B4
Summary ?
| GeneID | 10262 |
| Symbol | SF3B4 |
| Synonyms | AFD1|Hsh49|SAP49|SF3b49 |
| Description | splicing factor 3b subunit 4 |
| Reference | MIM:605593|HGNC:HGNC:10771|Ensembl:ENSG00000143368|HPRD:12028|Vega:OTTHUMG00000012208 |
| Gene type | protein-coding |
| Map location | 1q21.2 |
| Sherlock p-value | 0.164 |
| Fetal beta | 0.058 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03952484 | 1 | 149899818 | SF3B4 | 1.83E-10 | -0.013 | 5.29E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
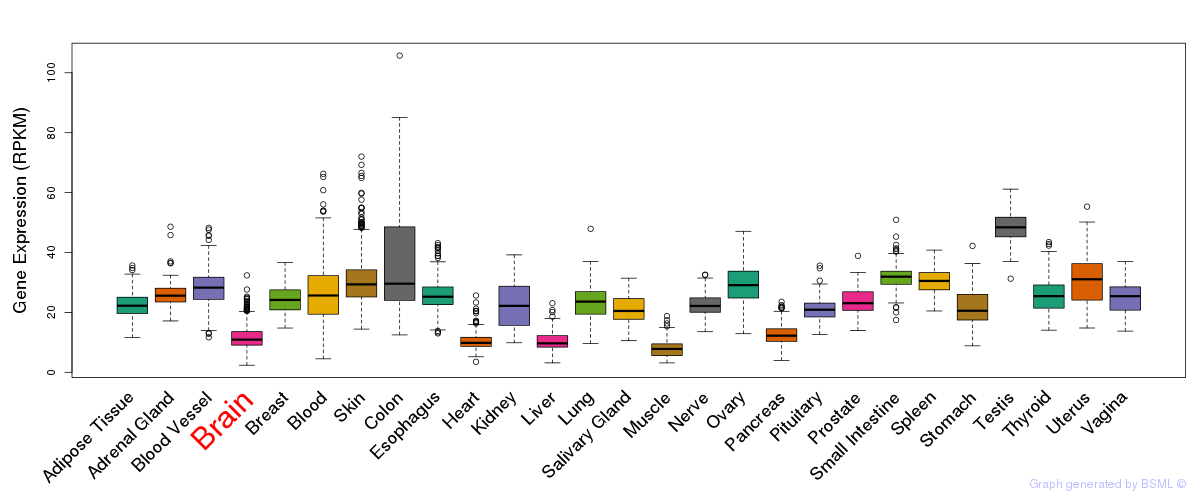
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SH3BP5L | 0.91 | 0.93 |
| RNF40 | 0.91 | 0.93 |
| LRRC47 | 0.91 | 0.93 |
| PRCC | 0.91 | 0.93 |
| FAM171A2 | 0.91 | 0.94 |
| PIK3R2 | 0.91 | 0.93 |
| STRN4 | 0.90 | 0.93 |
| LASP1 | 0.90 | 0.93 |
| TRIM46 | 0.90 | 0.94 |
| YY1 | 0.89 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.80 | -0.89 |
| AF347015.31 | -0.79 | -0.87 |
| AF347015.33 | -0.79 | -0.86 |
| AF347015.27 | -0.78 | -0.87 |
| MT-CYB | -0.77 | -0.87 |
| AF347015.8 | -0.76 | -0.87 |
| S100B | -0.75 | -0.82 |
| FXYD1 | -0.74 | -0.82 |
| COPZ2 | -0.74 | -0.80 |
| HLA-F | -0.74 | -0.76 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003676 | nucleic acid binding | IEA | - | |
| GO:0003723 | RNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 14667819 | |
| GO:0031202 | RNA splicing factor activity, transesterification mechanism | TAS | 7958871 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000398 | nuclear mRNA splicing, via spliceosome | EXP | 12226669 | |
| GO:0000398 | nuclear mRNA splicing, via spliceosome | IC | 9731529 | |
| GO:0008380 | RNA splicing | TAS | 7958871 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005681 | spliceosome | IDA | 9731529 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BMPR1A | 10q23del | ACVRLK3 | ALK3 | CD292 | bone morphogenetic protein receptor, type IA | - | HPRD,BioGRID | 15351706 |
| FAM115A | KIAA0738 | family with sequence similarity 115, member A | Two-hybrid | BioGRID | 16189514 |
| MAPK9 | JNK-55 | JNK2 | JNK2A | JNK2ALPHA | JNK2B | JNK2BETA | PRKM9 | SAPK | p54a | p54aSAPK | mitogen-activated protein kinase 9 | Two-hybrid | BioGRID | 16189514 |
| NCK2 | GRB4 | NCKbeta | NCK adaptor protein 2 | Two-hybrid | BioGRID | 16189514 |
| PHF5A | INI | MGC1346 | SF3b14b | bK223H9.2 | PHD finger protein 5A | - | HPRD | 12234937 |
| RNU11 | U11 | RNA, U11 small nuclear | Affinity Capture-MS | BioGRID | 15146077 |
| RNU12 | U12 | RNA, U12 small nuclear | Affinity Capture-MS | BioGRID | 15146077 |
| SF3A2 | PRP11 | PRPF11 | SAP62 | SF3a66 | splicing factor 3a, subunit 2, 66kDa | Affinity Capture-MS | BioGRID | 12234937 |
| SF3B1 | PRP10 | PRPF10 | SAP155 | SF3b155 | splicing factor 3b, subunit 1, 155kDa | Affinity Capture-MS | BioGRID | 12234937 |
| SF3B14 | CGI-110 | HSPC175 | Ht006 | P14 | SAP14 | SF3B14a | splicing factor 3B, 14 kDa subunit | - | HPRD | 12234937 |12738865 |
| SF3B2 | SAP145 | SF3B145 | SF3b1 | SF3b150 | splicing factor 3b, subunit 2, 145kDa | SAP145 interacts with SAP49. | BIND | 7958871 |
| SF3B2 | SAP145 | SF3B145 | SF3b1 | SF3b150 | splicing factor 3b, subunit 2, 145kDa | in vivo Two-hybrid | BioGRID | 7958871 |16189514 |
| SF3B2 | SAP145 | SF3B145 | SF3b1 | SF3b150 | splicing factor 3b, subunit 2, 145kDa | - | HPRD | 7958871 |10490618 |
| SMNDC1 | SMNR | SPF30 | survival motor neuron domain containing 1 | Affinity Capture-MS | BioGRID | 17353931 |
| SS18L1 | CREST | KIAA0693 | LP2261 | MGC26711 | MGC78386 | synovial sarcoma translocation gene on chromosome 18-like 1 | Two-hybrid | BioGRID | 16189514 |
| STK19 | D6S60 | D6S60E | G11 | HLA-RP1 | MGC117388 | RP1 | serine/threonine kinase 19 | - | HPRD | 14667819 |
| TCERG1 | CA150 | MGC133200 | TAF2S | transcription elongation regulator 1 | - | HPRD,BioGRID | 15456888 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING | 111 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING MINOR PATHWAY | 45 | 19 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN | 187 | 122 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR UP | 151 | 100 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN | 128 | 90 | All SZGR 2.0 genes in this pathway |
| JAZAERI BREAST CANCER BRCA1 VS BRCA2 UP | 49 | 28 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS DN | 145 | 93 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-133 | 54 | 60 | 1A | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.