Gene Page: PIAS3
Summary ?
| GeneID | 10401 |
| Symbol | PIAS3 |
| Synonyms | ZMIZ5 |
| Description | protein inhibitor of activated STAT 3 |
| Reference | MIM:605987|HGNC:HGNC:16861|Ensembl:ENSG00000131788|HPRD:09068|Vega:OTTHUMG00000013750 |
| Gene type | protein-coding |
| Map location | 1q21 |
| Sherlock p-value | 0.033 |
| Fetal beta | -0.278 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | PIAS3 | 10401 | 0.16 | trans |
Section II. Transcriptome annotation
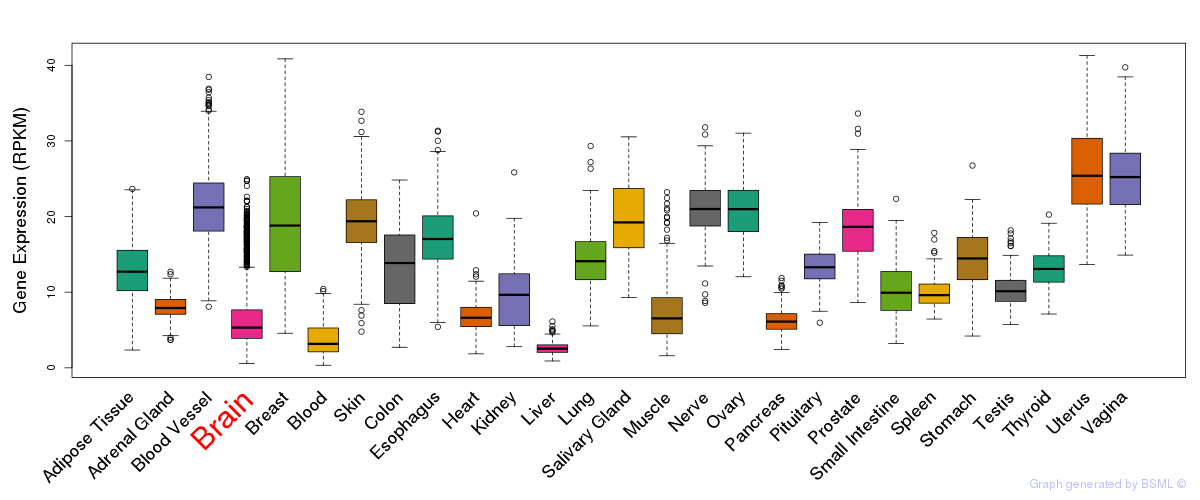
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FHL1 | 0.83 | 0.84 |
| NAGA | 0.83 | 0.79 |
| GLB1 | 0.82 | 0.79 |
| HEATR2 | 0.81 | 0.85 |
| SALL2 | 0.81 | 0.81 |
| RP11-369J21.1 | 0.80 | 0.77 |
| TCTN1 | 0.79 | 0.72 |
| TCTN3 | 0.79 | 0.72 |
| DOLK | 0.79 | 0.80 |
| C1orf56 | 0.78 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.59 | -0.68 |
| MT-CO2 | -0.59 | -0.67 |
| AF347015.21 | -0.58 | -0.71 |
| AF347015.8 | -0.57 | -0.67 |
| AF347015.31 | -0.57 | -0.66 |
| AF347015.33 | -0.56 | -0.64 |
| MT-CYB | -0.54 | -0.63 |
| AC098691.2 | -0.53 | -0.62 |
| MT-ATP8 | -0.51 | -0.70 |
| AF347015.2 | -0.51 | -0.64 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003676 | nucleic acid binding | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0051059 | NF-kappaB binding | IPI | 15140884 | |
| GO:0019789 | SUMO ligase activity | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006511 | ubiquitin-dependent protein catabolic process | IEA | - | |
| GO:0016925 | protein sumoylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0016607 | nuclear speck | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Two-hybrid | BioGRID | 11117529 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | Affinity Capture-Western | BioGRID | 14691252 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | Affinity Capture-Western | BioGRID | 14691252 |
| GEMIN4 | DKFZp434B131 | DKFZp434D174 | HC56 | HCAP1 | HHRF-1 | p97 | gem (nuclear organelle) associated protein 4 | HCAP1 interacts with PIAS3 | BIND | 12869526 |
| GFI1 | FLJ94509 | GFI-1 | ZNF163 | growth factor independent 1 transcription repressor | Two-hybrid | BioGRID | 11060035 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD | 14691252 |
| HMGA2 | BABL | HMGI-C | HMGIC | LIPO | STQTL9 | high mobility group AT-hook 2 | Affinity Capture-Western Two-hybrid | BioGRID | 11390395 |
| MITF | MI | WS2A | bHLHe32 | microphthalmia-associated transcription factor | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 11709556 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 15140884 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD,BioGRID | 14691252 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD,BioGRID | 14691252 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | - | HPRD,BioGRID | 14691252 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD | 11429412 |12804609 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD,BioGRID | 11429412 |
| ZFHX3 | ATBF1 | ATBT | zinc finger homeobox 3 | ATBF1 interacts with PIAS3. | BIND | 14715251 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG UBIQUITIN MEDIATED PROTEOLYSIS | 138 | 98 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| ST JAK STAT PATHWAY | 9 | 8 | All SZGR 2.0 genes in this pathway |
| ST STAT3 PATHWAY | 11 | 10 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| PID IL6 7 PATHWAY | 47 | 40 | All SZGR 2.0 genes in this pathway |
| PID CMYB PATHWAY | 84 | 61 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION | 77 | 51 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION CCNE1 | 40 | 26 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION APOBEC2 | 27 | 19 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION 12HR | 43 | 35 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 3 | 48 | 31 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON VS RECTAL CANCER DN | 56 | 36 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION C | 69 | 49 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| WORSCHECH TUMOR EVASION AND TOLEROGENICITY UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 17 | 181 | 101 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 238 | 244 | m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-18 | 774 | 780 | m8 | hsa-miR-18a | UAAGGUGCAUCUAGUGCAGAUA |
| hsa-miR-18b | UAAGGUGCAUCUAGUGCAGUUA | ||||
| miR-181 | 916 | 922 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-324-3p | 263 | 269 | m8 | hsa-miR-324-3p | CCACUGCCCCAGGUGCUGCUGG |
| miR-410 | 690 | 696 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-9 | 572 | 578 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.