Gene Page: UNC13B
Summary ?
| GeneID | 10497 |
| Symbol | UNC13B |
| Synonyms | MUNC13|UNC13|Unc13h2 |
| Description | unc-13 homolog B (C. elegans) |
| Reference | MIM:605836|HGNC:HGNC:12566|Ensembl:ENSG00000198722|HPRD:05785|Vega:OTTHUMG00000019856 |
| Gene type | protein-coding |
| Map location | 9p13.3 |
| Pascal p-value | 0.007 |
| Sherlock p-value | 0.709 |
| Fetal beta | 0.146 |
| Support | CANABINOID EXOCYTOSIS METABOTROPIC GLUTAMATE RECEPTOR SEROTONIN Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
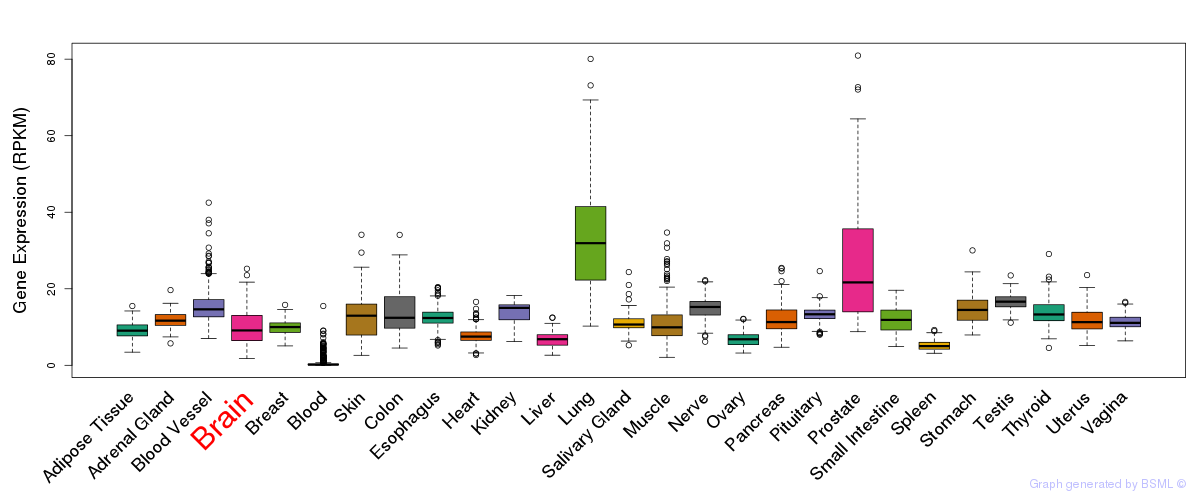
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZFP62 | 0.93 | 0.78 |
| TET1 | 0.92 | 0.81 |
| EPB41 | 0.92 | 0.75 |
| SMARCC1 | 0.91 | 0.82 |
| IGF2BP3 | 0.91 | 0.85 |
| CCDC123 | 0.91 | 0.86 |
| FUBP1 | 0.91 | 0.80 |
| CASP2 | 0.91 | 0.80 |
| LRIG2 | 0.91 | 0.74 |
| NONO | 0.91 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FBXO2 | -0.68 | -0.76 |
| C5orf53 | -0.67 | -0.81 |
| LHPP | -0.67 | -0.65 |
| HLA-F | -0.67 | -0.74 |
| TNFSF12 | -0.66 | -0.71 |
| ALDOC | -0.66 | -0.74 |
| LDHD | -0.66 | -0.72 |
| PTH1R | -0.66 | -0.68 |
| AIFM3 | -0.65 | -0.71 |
| SLC16A11 | -0.65 | -0.67 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | TAS | 9607201 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0019992 | diacylglycerol binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | IEA | neuron, Synap, Neurotransmitter (GO term level: 6) | - |
| GO:0016082 | synaptic vesicle priming | IEA | Synap (GO term level: 10) | - |
| GO:0007242 | intracellular signaling cascade | IEA | - | |
| GO:0006917 | induction of apoptosis | TAS | 10233166 | |
| GO:0007588 | excretion | TAS | 9607201 | |
| GO:0006887 | exocytosis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005794 | Golgi apparatus | TAS | 9607201 | |
| GO:0005737 | cytoplasm | TAS | 10233166 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | 15 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RELEASE CYCLE | 34 | 30 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C UP | 170 | 114 | All SZGR 2.0 genes in this pathway |
| HEIDENBLAD AMPLICON 8Q24 UP | 40 | 23 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE UP | 283 | 177 | All SZGR 2.0 genes in this pathway |
| NELSON RESPONSE TO ANDROGEN UP | 86 | 61 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 UP | 56 | 34 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION UP | 140 | 83 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-151 | 860 | 867 | 1A,m8 | hsa-miR-151brain | ACUAGACUGAAGCUCCUUGAGG |
| miR-183 | 1249 | 1255 | m8 | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.