Gene Page: SEMA4D
Summary ?
| GeneID | 10507 |
| Symbol | SEMA4D |
| Synonyms | C9orf164|CD100|M-sema-G|SEMAJ|coll-4 |
| Description | semaphorin 4D |
| Reference | MIM:601866|HGNC:HGNC:10732|Ensembl:ENSG00000187764|HPRD:03520|Vega:OTTHUMG00000020185 |
| Gene type | protein-coding |
| Map location | 9q22.2 |
| Pascal p-value | 0.732 |
| Sherlock p-value | 0.764 |
| Fetal beta | -2.096 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
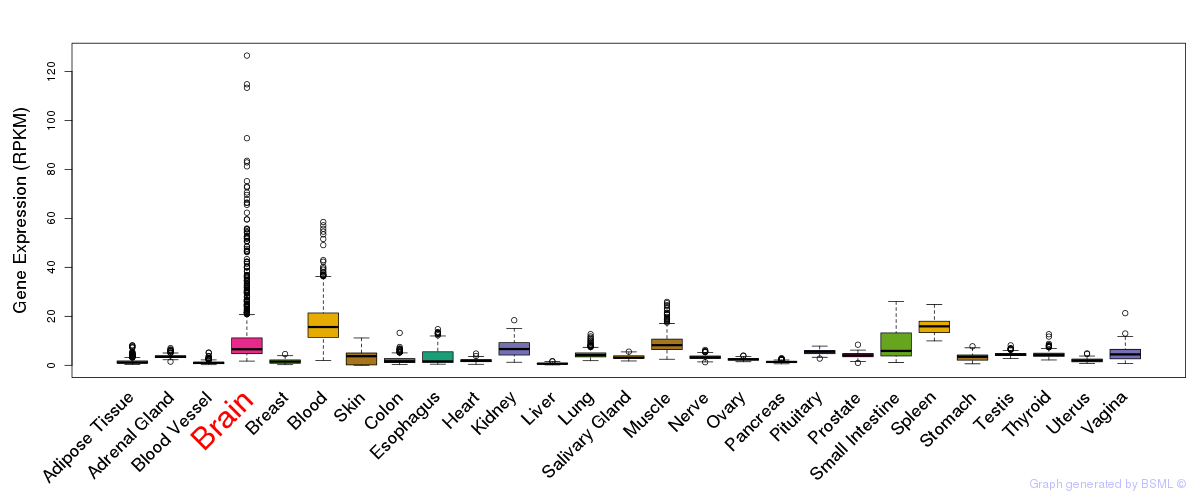
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CHMP2A | 0.84 | 0.82 |
| MEA1 | 0.82 | 0.81 |
| TIMM13 | 0.81 | 0.80 |
| C16orf42 | 0.79 | 0.74 |
| SPAG7 | 0.79 | 0.75 |
| AC010422.1 | 0.79 | 0.77 |
| HRAS | 0.79 | 0.78 |
| CCDC124 | 0.78 | 0.76 |
| CHID1 | 0.78 | 0.74 |
| SURF2 | 0.77 | 0.78 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.8 | -0.49 | -0.42 |
| MT-ATP8 | -0.48 | -0.46 |
| AF347015.33 | -0.47 | -0.41 |
| AF347015.15 | -0.46 | -0.43 |
| AF347015.26 | -0.46 | -0.42 |
| AF347015.27 | -0.46 | -0.42 |
| AF347015.21 | -0.46 | -0.34 |
| MT-CYB | -0.46 | -0.41 |
| MT-CO2 | -0.45 | -0.37 |
| AF347015.2 | -0.45 | -0.38 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | IDA | Neurotransmitter (GO term level: 4) | 11254688 |
| GO:0004872 | receptor activity | IDA | 15613544 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0050772 | positive regulation of axonogenesis | IEA | axon, neurogenesis (GO term level: 14) | - |
| GO:0007399 | nervous system development | IEA | neurite (GO term level: 5) | - |
| GO:0007155 | cell adhesion | TAS | 8876214 | |
| GO:0006955 | immune response | TAS | 8876214 | |
| GO:0006916 | anti-apoptosis | TAS | 8876214 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA4D IN SEMAPHORIN SIGNALING | 32 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMAPHORIN INTERACTIONS | 68 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | 27 | 21 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA PROGENITOR UP | 39 | 25 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 4 | 61 | 34 | All SZGR 2.0 genes in this pathway |
| AMUNDSON GENOTOXIC SIGNATURE | 105 | 68 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| MORI LARGE PRE BII LYMPHOCYTE DN | 58 | 39 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE UP | 53 | 35 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA HP UP | 49 | 26 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO OR UV | 63 | 44 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| PARK TRETINOIN RESPONSE AND PML RARA FUSION | 30 | 21 | All SZGR 2.0 genes in this pathway |
| HOFFMANN SMALL PRE BII TO IMMATURE B LYMPHOCYTE UP | 70 | 49 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| NABA ECM AFFILIATED | 171 | 89 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-125/351 | 626 | 633 | 1A,m8 | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-188 | 629 | 635 | 1A | hsa-miR-188 | CAUCCCUUGCAUGGUGGAGGGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.