Gene Page: POLR3C
Summary ?
| GeneID | 10623 |
| Symbol | POLR3C |
| Synonyms | RPC3|RPC62 |
| Description | polymerase (RNA) III subunit C |
| Reference | HGNC:HGNC:30076|Ensembl:ENSG00000186141|HPRD:17880|Vega:OTTHUMG00000013753 |
| Gene type | protein-coding |
| Map location | 1q21.1 |
| Sherlock p-value | 0.866 |
| Fetal beta | 0.28 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
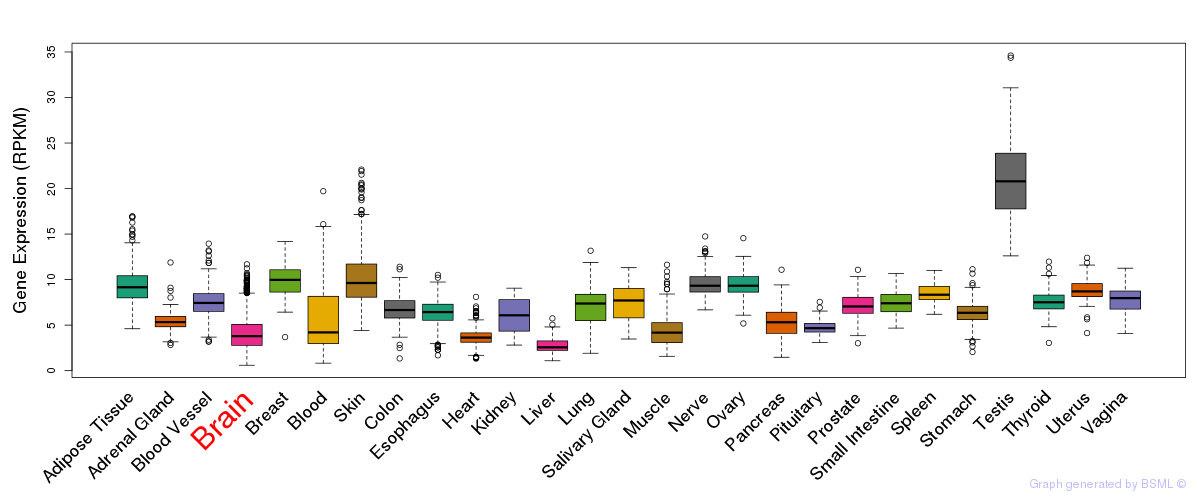
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF700 | 0.90 | 0.89 |
| ZNF334 | 0.90 | 0.86 |
| CCNL1 | 0.89 | 0.84 |
| ZNF177 | 0.89 | 0.87 |
| ZNF140 | 0.89 | 0.90 |
| ZNF211 | 0.89 | 0.86 |
| ZNF528 | 0.88 | 0.86 |
| ZNF7 | 0.88 | 0.89 |
| DMTF1 | 0.88 | 0.87 |
| EXOSC10 | 0.88 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.67 | -0.81 |
| AF347015.27 | -0.67 | -0.79 |
| MT-CO2 | -0.66 | -0.79 |
| MT-CYB | -0.65 | -0.80 |
| AF347015.8 | -0.65 | -0.81 |
| AIFM3 | -0.65 | -0.74 |
| AF347015.33 | -0.64 | -0.78 |
| IFI27 | -0.63 | -0.79 |
| AF347015.15 | -0.63 | -0.79 |
| HLA-F | -0.63 | -0.69 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0003899 | DNA-directed RNA polymerase activity | IEA | - | |
| GO:0003899 | DNA-directed RNA polymerase activity | TAS | 9171375 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006359 | regulation of transcription from RNA polymerase III promoter | TAS | 9171375 | |
| GO:0006350 | transcription | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005666 | DNA-directed RNA polymerase III complex | TAS | 9171375 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PURINE METABOLISM | 159 | 96 | All SZGR 2.0 genes in this pathway |
| KEGG PYRIMIDINE METABOLISM | 98 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG RNA POLYMERASE | 29 | 12 | All SZGR 2.0 genes in this pathway |
| KEGG CYTOSOLIC DNA SENSING PATHWAY | 56 | 44 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | 23 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION | 33 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | 26 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III TRANSCRIPTION TERMINATION | 19 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL III CHAIN ELONGATION | 17 | 8 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS UP | 112 | 68 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS INTERMEDIATE PROGENITOR | 149 | 84 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |