Gene Page: CFTR
Summary ?
| GeneID | 1080 |
| Symbol | CFTR |
| Synonyms | ABC35|ABCC7|CF|CFTR/MRP|MRP7|TNR-CFTR|dJ760C5.1 |
| Description | cystic fibrosis transmembrane conductance regulator |
| Reference | MIM:602421|HGNC:HGNC:1884|Ensembl:ENSG00000001626|HPRD:03883|Vega:OTTHUMG00000023076 |
| Gene type | protein-coding |
| Map location | 7q31.2 |
| Pascal p-value | 0.115 |
| Fetal beta | -0.251 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 1.469 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0352 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | CFTR | 1080 | 0.01 | trans | ||
| rs17714461 | chr19 | 51425105 | CFTR | 1080 | 0.18 | trans |
Section II. Transcriptome annotation
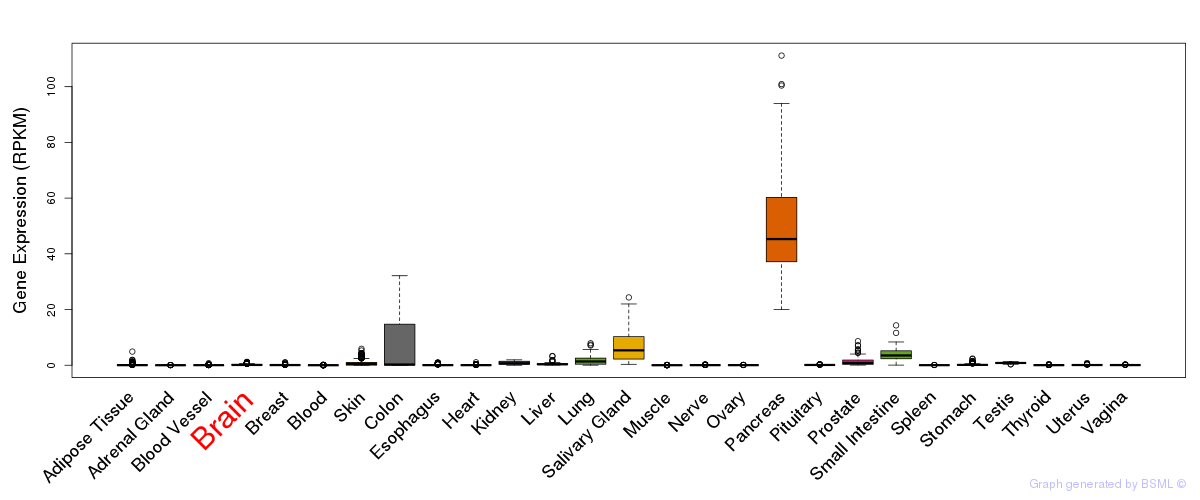
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 11707463 |15247260 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0005524 | ATP binding | TAS | 2475911 | |
| GO:0005254 | chloride channel activity | IEA | - | |
| GO:0005260 | channel-conductance-controlling ATPase activity | NAS | 11707463 | |
| GO:0005216 | ion channel activity | IEA | - | |
| GO:0005224 | ATP-binding and phosphorylation-dependent chloride channel activity | TAS | 10581360 | |
| GO:0030165 | PDZ domain binding | IDA | 11707463 | |
| GO:0016887 | ATPase activity | IEA | - | |
| GO:0031404 | chloride ion binding | IEA | - | |
| GO:0042626 | ATPase activity, coupled to transmembrane movement of substances | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007585 | respiratory gaseous exchange | TAS | 9875854 | |
| GO:0006811 | ion transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0016323 | basolateral plasma membrane | NAS | 11707463 | |
| GO:0016324 | apical plasma membrane | IDA | 15247260 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CANX | CNX | FLJ26570 | IP90 | P90 | calnexin | CFTR interacts with CANX. This interaction was modeled on a demonstrated interaction between human CFTR and hamster CANX. | BIND | 15105504 |
| CANX | CNX | FLJ26570 | IP90 | P90 | calnexin | CFTR interacts with CANX. | BIND | 7513695 |
| CFTR | ABC35 | ABCC7 | CF | CFTR/MRP | MRP7 | TNR-CFTR | dJ760C5.1 | cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) | - | HPRD,BioGRID | 11051556 |
| CFTR | ABC35 | ABCC7 | CF | CFTR/MRP | MRP7 | TNR-CFTR | dJ760C5.1 | cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) | CFTR forms a homodimer | BIND | 12933354 |
| CLCN3 | CLC3 | ClC-3 | chloride channel 3 | Reconstituted Complex | BioGRID | 12471024 |
| DNAJA1 | DJ-2 | DjA1 | HDJ2 | HSDJ | HSJ2 | HSPF4 | hDJ-2 | DnaJ (Hsp40) homolog, subfamily A, member 1 | - | HPRD,BioGRID | 10075921 |
| DNAJB1 | HSPF1 | Hdj1 | Hsp40 | Sis1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | Affinity Capture-Western | BioGRID | 10075921 |
| DNAJC5 | CSP | DKFZp434N1429 | DKFZp761N1221 | DNAJC5A | FLJ00118 | FLJ13070 | DnaJ (Hsp40) homolog, subfamily C, member 5 | Csp1 interacts with CFTR. | BIND | 12039948 |
| DNAJC5 | CSP | DKFZp434N1429 | DKFZp761N1221 | DNAJC5A | FLJ00118 | FLJ13070 | DnaJ (Hsp40) homolog, subfamily C, member 5 | in vitro | BioGRID | 12039948 |
| DNAJC5 | CSP | DKFZp434N1429 | DKFZp761N1221 | DNAJC5A | FLJ00118 | FLJ13070 | DnaJ (Hsp40) homolog, subfamily C, member 5 | Csp2 interacts with CFTR. | BIND | 12039948 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | - | HPRD,BioGRID | 10893422 |
| GOPC | CAL | FIG | GOPC1 | PIST | dJ94G16.2 | golgi associated PDZ and coiled-coil motif containing | - | HPRD,BioGRID | 11707463 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | Affinity Capture-Western | BioGRID | 10075921 |
| KCNJ1 | KIR1.1 | ROMK | ROMK1 | potassium inwardly-rectifying channel, subfamily J, member 1 | Affinity Capture-Western | BioGRID | 14604981 |
| PDZD3 | FLJ22756 | IKEPP | PDZK2 | PDZ domain containing 3 | Reconstituted Complex | BioGRID | 12615054 |
| PDZK1 | CAP70 | CLAMP | NHERF3 | PDZD1 | PDZ domain containing 1 | - | HPRD,BioGRID | 11051556 |12471024 |12615054 |
| PRKAA1 | AMPK | AMPKa1 | MGC33776 | MGC57364 | protein kinase, AMP-activated, alpha 1 catalytic subunit | - | HPRD,BioGRID | 10862786 |
| PRKAR2A | MGC3606 | PKR2 | PRKAR2 | protein kinase, cAMP-dependent, regulatory, type II, alpha | PKA II interacts with and phosphorylates CFTR. | BIND | 10799517 |
| PRKCE | MGC125656 | MGC125657 | PKCE | nPKC-epsilon | protein kinase C, epsilon | - | HPRD,BioGRID | 11956211 |
| SLC4A7 | DKFZp686H168 | NBC2 | NBC3 | SBC2 | SLC4A6 | solute carrier family 4, sodium bicarbonate cotransporter, member 7 | - | HPRD,BioGRID | 12403779 |
| SLC4A8 | DKFZp761B2318 | FLJ46462 | NBC3 | solute carrier family 4, sodium bicarbonate cotransporter, member 8 | - | HPRD,BioGRID | 12403779 |
| SLC9A3R1 | EBP50 | NHERF | NHERF1 | NPHLOP2 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 | First PDZ domain of EBP50 binds to the C-terminus of CFTR. | BIND | 9671706 |9677412 |10852925 |12621035 |
| SLC9A3R1 | EBP50 | NHERF | NHERF1 | NPHLOP2 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 | - | HPRD | 9613608 |9677412 |10852925 |
| SLC9A3R1 | EBP50 | NHERF | NHERF1 | NPHLOP2 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 | Affinity Capture-Western in vitro in vivo Reconstituted Complex | BioGRID | 9613608 |9671706 |9677412 |10852925 |12471024 |12615054 |
| SLC9A3R2 | E3KARP | MGC104639 | NHE3RF2 | NHERF-2 | NHERF2 | OCTS2 | SIP-1 | SIP1 | TKA-1 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 | - | HPRD,BioGRID | 9671706 |10893422 |
| SLC9A3R2 | E3KARP | MGC104639 | NHE3RF2 | NHERF-2 | NHERF2 | OCTS2 | SIP-1 | SIP1 | TKA-1 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 | The second PDZ domain of E3KARP interacts with the C-terminal PDZ binding motif of CFTR. | BIND | 10893422 |
| SLC9A6 | KIAA0267 | MRSA | NHE6 | solute carrier family 9 (sodium/hydrogen exchanger), member 6 | in vivo | BioGRID | 11707463 |
| SNAP23 | HsT17016 | SNAP23A | SNAP23B | synaptosomal-associated protein, 23kDa | - | HPRD,BioGRID | 12209004 |
| STX1A | HPC-1 | STX1 | p35-1 | syntaxin 1A (brain) | Syntaxin 1A interacts with CFTR. | BIND | 9384384 |12446681 |
| STX1A | HPC-1 | STX1 | p35-1 | syntaxin 1A (brain) | - | HPRD,BioGRID | 9384384 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ABC TRANSPORTERS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| KEGG VIBRIO CHOLERAE INFECTION | 56 | 32 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CFTR PATHWAY | 12 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | 34 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE UP | 108 | 67 | All SZGR 2.0 genes in this pathway |
| WATANABE COLON CANCER MSI VS MSS DN | 81 | 42 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY DN | 138 | 70 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| MIDORIKAWA AMPLIFIED IN LIVER CANCER | 55 | 38 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2 | 127 | 92 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| WAGSCHAL EHMT2 TARGETS UP | 13 | 7 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 1508 | 1514 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-144 | 1508 | 1514 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-377 | 1549 | 1555 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.