Gene Page: HPSE
Summary ?
| GeneID | 10855 |
| Symbol | HPSE |
| Synonyms | HPA|HPA1|HPR1|HPSE1|HSE1 |
| Description | heparanase |
| Reference | MIM:604724|HGNC:HGNC:5164|Ensembl:ENSG00000173083|HPRD:05286|Vega:OTTHUMG00000130425 |
| Gene type | protein-coding |
| Map location | 4q21.3 |
| Pascal p-value | 0.648 |
| Sherlock p-value | 0.286 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20587127 | 4 | 84255925 | HPSE | 1.67E-9 | -0.02 | 1.48E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4431217 | chr4 | 84150544 | HPSE | 10855 | 0.11 | cis | ||
| rs17006618 | chr4 | 84151091 | HPSE | 10855 | 0.14 | cis | ||
| rs17006620 | chr4 | 84151984 | HPSE | 10855 | 0.14 | cis | ||
| rs6840203 | chr4 | 84153457 | HPSE | 10855 | 0.14 | cis | ||
| rs6840620 | chr4 | 84153609 | HPSE | 10855 | 0.02 | cis | ||
| rs17006628 | chr4 | 84154498 | HPSE | 10855 | 0.14 | cis | ||
| rs6816252 | chr4 | 84160091 | HPSE | 10855 | 0.12 | cis | ||
| rs6841309 | chr4 | 84160132 | HPSE | 10855 | 0.14 | cis | ||
| rs4508871 | chr4 | 84161318 | HPSE | 10855 | 0.14 | cis | ||
| rs7672110 | chr4 | 84161713 | HPSE | 10855 | 0.14 | cis | ||
| rs7672528 | chr4 | 84161949 | HPSE | 10855 | 0.14 | cis |
Section II. Transcriptome annotation
General gene expression (GTEx)
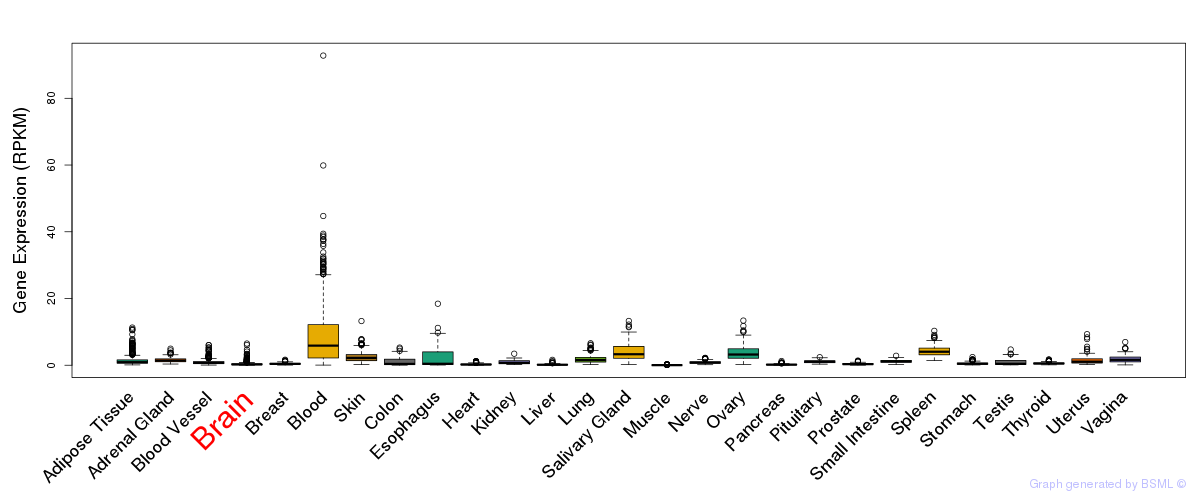
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HAUS1 | 0.93 | 0.91 |
| SNRPF | 0.92 | 0.87 |
| RPL6P10 | 0.92 | 0.85 |
| TUT1 | 0.92 | 0.87 |
| YBX1 | 0.92 | 0.85 |
| CCDC23 | 0.92 | 0.84 |
| EIF3H | 0.92 | 0.81 |
| BTF3 | 0.91 | 0.82 |
| ILF2 | 0.91 | 0.80 |
| DYNLT1 | 0.91 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.69 | -0.79 |
| FBXO2 | -0.69 | -0.78 |
| ALDOC | -0.67 | -0.75 |
| PTH1R | -0.67 | -0.71 |
| AIFM3 | -0.67 | -0.72 |
| C5orf53 | -0.67 | -0.65 |
| LDHD | -0.65 | -0.74 |
| CLU | -0.65 | -0.75 |
| LHPP | -0.65 | -0.67 |
| ATP10A | -0.65 | -0.75 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOSAMINOGLYCAN DEGRADATION | 21 | 14 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 1 PATHWAY | 46 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME HS GAG DEGRADATION | 20 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME HEPARAN SULFATE HEPARIN HS GAG METABOLISM | 52 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOSAMINOGLYCAN METABOLISM | 111 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| SMIRNOV CIRCULATING ENDOTHELIOCYTES IN CANCER UP | 158 | 103 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| SEITZ NEOPLASTIC TRANSFORMATION BY 8P DELETION UP | 73 | 47 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 1 UP | 118 | 66 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS DN | 120 | 69 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY DEACETYLATION IN PANCREATIC CANCER | 50 | 30 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS DN | 142 | 94 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| LI PROSTATE CANCER EPIGENETIC | 30 | 22 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA | 140 | 96 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| ISSAEVA MLL2 TARGETS | 62 | 35 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |