Gene Page: CPNE8
Summary ?
| GeneID | 144402 |
| Symbol | CPNE8 |
| Synonyms | - |
| Description | copine 8 |
| Reference | HGNC:HGNC:23498|Ensembl:ENSG00000139117|HPRD:16747|Vega:OTTHUMG00000169396 |
| Gene type | protein-coding |
| Map location | 12q12 |
| Pascal p-value | 0.064 |
| Sherlock p-value | 0.028 |
| Fetal beta | -0.695 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| Expression | Meta-analysis of gene expression | P value: 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20857455 | 12 | 39300142 | CPNE8 | -0.035 | 0.86 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7882576 | chrX | 38501487 | CPNE8 | 144402 | 0.08 | trans |
Section II. Transcriptome annotation
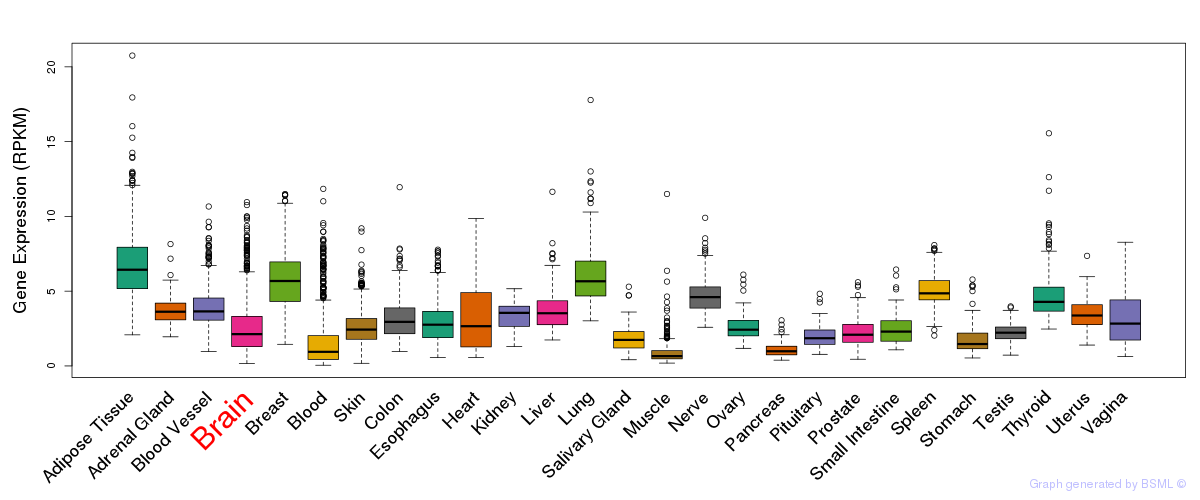
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY UP | 175 | 120 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS GREEN UP | 23 | 19 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO BEXAROTENE DN | 29 | 19 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 VS CD2 UP | 66 | 47 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE S | 162 | 86 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 1197 | 1203 | 1A | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-137 | 489 | 495 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-151 | 905 | 911 | 1A | hsa-miR-151brain | ACUAGACUGAAGCUCCUUGAGG |
| miR-186 | 1181 | 1187 | 1A | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-188 | 924 | 931 | 1A,m8 | hsa-miR-188 | CAUCCCUUGCAUGGUGGAGGGU |
| miR-204/211 | 923 | 929 | m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-218 | 302 | 309 | 1A,m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-221/222 | 865 | 872 | 1A,m8 | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-224 | 1391 | 1397 | m8 | hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA |
| miR-25/32/92/363/367 | 468 | 474 | 1A | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-29 | 62 | 68 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-299-5p | 156 | 162 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-30-5p | 376 | 383 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-375 | 1188 | 1194 | 1A | hsa-miR-375 | UUUGUUCGUUCGGCUCGCGUGA |
| miR-493-5p | 1125 | 1131 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-539 | 96 | 102 | 1A | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.