Gene Page: CSK
Summary ?
| GeneID | 1445 |
| Symbol | CSK |
| Synonyms | - |
| Description | c-src tyrosine kinase |
| Reference | MIM:124095|HGNC:HGNC:2444|Ensembl:ENSG00000103653|HPRD:00496|Vega:OTTHUMG00000142814 |
| Gene type | protein-coding |
| Map location | 15q24.1 |
| Pascal p-value | 0.015 |
| Sherlock p-value | 0.374 |
| Fetal beta | -0.145 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0102 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
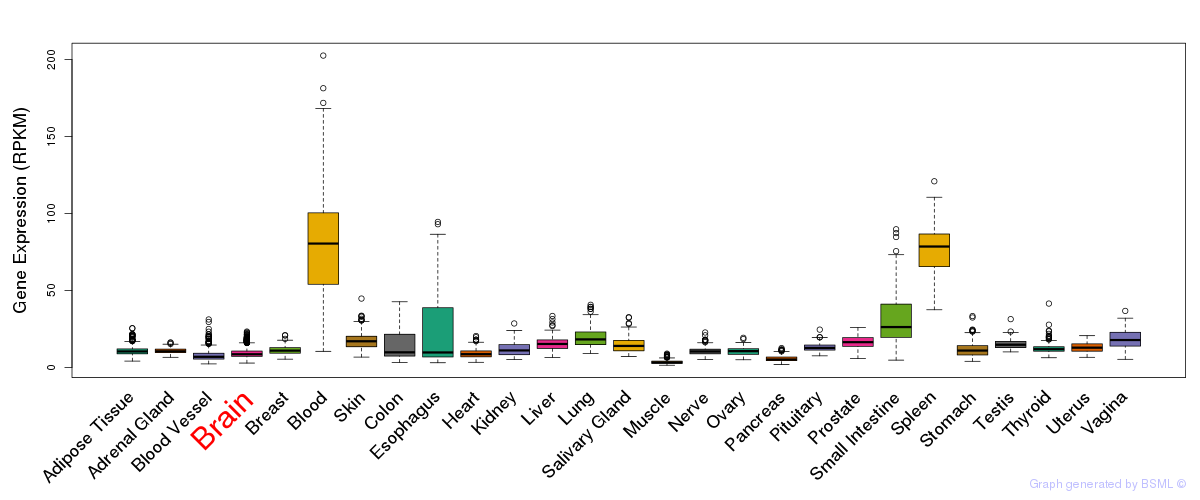
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| UBE3A | 0.97 | 0.97 |
| IPO7 | 0.96 | 0.97 |
| BCLAF1 | 0.96 | 0.97 |
| DIS3 | 0.95 | 0.96 |
| ZMYM2 | 0.95 | 0.96 |
| FYTTD1 | 0.95 | 0.96 |
| CAND1 | 0.95 | 0.97 |
| KPNA4 | 0.95 | 0.95 |
| SMEK2 | 0.95 | 0.95 |
| PPP2R5E | 0.95 | 0.96 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.77 | -0.86 |
| MT-CO2 | -0.75 | -0.85 |
| AF347015.31 | -0.74 | -0.83 |
| AF347015.33 | -0.73 | -0.81 |
| HIGD1B | -0.73 | -0.84 |
| IFI27 | -0.72 | -0.83 |
| MT-CYB | -0.72 | -0.81 |
| TSC22D4 | -0.71 | -0.78 |
| HSD17B14 | -0.71 | -0.76 |
| AF347015.8 | -0.71 | -0.82 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004715 | non-membrane spanning protein tyrosine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008022 | protein C-terminus binding | TAS | 8890164 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | TAS | 1945408 | |
| GO:0008285 | negative regulation of cell proliferation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | TAS | 8890164 | |
| GO:0005911 | cell-cell junction | IEA | - | |
| GO:0005886 | plasma membrane | EXP | 15574420 | |
| GO:0005886 | plasma membrane | IPI | 10790433 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARRB1 | ARB1 | ARR1 | arrestin, beta 1 | - | HPRD,BioGRID | 10753943 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | - | HPRD,BioGRID | 11805080 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | - | HPRD,BioGRID | 11149930 |
| CD247 | CD3-ZETA | CD3H | CD3Q | CD3Z | T3Z | TCRZ | CD247 molecule | - | HPRD | 9172452 |
| CD44 | CDW44 | CSPG8 | ECMR-III | HCELL | IN | LHR | MC56 | MDU2 | MDU3 | MGC10468 | MIC4 | MUTCH-I | Pgp1 | CD44 molecule (Indian blood group) | - | HPRD | 11084024 |
| CDH5 | 7B4 | CD144 | FLJ17376 | cadherin 5, type 2 (vascular endothelium) | CDH5 (VE-cadherin) interacts with CSK. | BIND | 15861137 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD,BioGRID | 10801129 |
| CSK | MGC117393 | c-src tyrosine kinase | CSK autophosphorylates. This interaction was modeled on a demonstrated interaction using CSK from an unspecified species. | BIND | 12588871 |
| DAB2 | DOC-2 | DOC2 | FLJ26626 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | - | HPRD,BioGRID | 12473651 |
| DAG1 | 156DAG | A3a | AGRNR | DAG | dystroglycan 1 (dystrophin-associated glycoprotein 1) | Affinity Capture-Western Reconstituted Complex | BioGRID | 11724572 |
| DOK3 | DOKL | FLJ22570 | FLJ39939 | docking protein 3 | - | HPRD | 10733577 |14993273 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | Affinity Capture-MS | BioGRID | 16729043 |
| ERBB3 | ErbB-3 | HER3 | LCCS2 | MDA-BF-1 | MGC88033 | c-erbB-3 | c-erbB3 | erbB3-S | p180-ErbB3 | p45-sErbB3 | p85-sErbB3 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | Affinity Capture-MS | BioGRID | 16729043 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 8262983 |
| HNRNPK | CSBP | FLJ41122 | HNRPK | TUNP | heterogeneous nuclear ribonucleoprotein K | - | HPRD,BioGRID | 12052863 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | CSK interacts with IGF-IR. | BIND | 10026153 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | - | HPRD,BioGRID | 10026153 |
| INSR | CD220 | HHF5 | insulin receptor | CSK interacts with IR. | BIND | 10026153 |
| INSR | CD220 | HHF5 | insulin receptor | - | HPRD,BioGRID | 10026153 |
| PAG1 | CBP | FLJ37858 | MGC138364 | PAG | phosphoprotein associated with glycosphingolipid microdomains 1 | - | HPRD,BioGRID | 10790433 |
| PECAM1 | CD31 | PECAM-1 | platelet/endothelial cell adhesion molecule | - | HPRD,BioGRID | 9624175 |
| PLD2 | - | phospholipase D2 | - | HPRD | 12697812 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD,BioGRID | 7529872 |
| PTPN12 | PTP-PEST | PTPG1 | protein tyrosine phosphatase, non-receptor type 12 | - | HPRD | 11158295 |
| PXN | FLJ16691 | paxillin | - | HPRD,BioGRID | 7529872 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | - | HPRD,BioGRID | 7544435 |
| RGS16 | A28-RGS14 | A28-RGS14P | RGS-R | regulator of G-protein signaling 16 | CSK phosphorylates RGS16. This interaction was modeled on a demonstrated interaction between CSK from an unspecified species and human RSG16. | BIND | 12588871 |
| SDC3 | N-syndecan | SDCN | SYND3 | syndecan 3 | - | HPRD,BioGRID | 9553134 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 10037694 |12048194 |
| SIT1 | MGC125908 | MGC125909 | MGC125910 | RP11-331F9.5 | SIT | signaling threshold regulating transmembrane adaptor 1 | - | HPRD | 11433379 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 7513429 |
| TGFB1I1 | ARA55 | HIC-5 | HIC5 | TSC-5 | transforming growth factor beta 1 induced transcript 1 | Reconstituted Complex | BioGRID | 9858471 |
| YTHDC1 | KIAA1966 | YT521 | YT521-B | YTH domain containing 1 | Affinity Capture-Western Biochemical Activity | BioGRID | 15175272 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG EPITHELIAL CELL SIGNALING IN HELICOBACTER PYLORI INFECTION | 68 | 44 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CSK PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SRCRPTP PATHWAY | 11 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CELL2CELL PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTEGRIN PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| ST T CELL SIGNAL TRANSDUCTION | 45 | 33 | All SZGR 2.0 genes in this pathway |
| SIG BCR SIGNALING PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| ST B CELL ANTIGEN RECEPTOR | 40 | 32 | All SZGR 2.0 genes in this pathway |
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| PID TCR PATHWAY | 66 | 51 | All SZGR 2.0 genes in this pathway |
| PID PTP1B PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR PATHWAY | 53 | 42 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME TCR SIGNALING | 54 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME GAB1 SIGNALOSOME | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | 16 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME PD1 SIGNALING | 18 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME COSTIMULATION BY THE CD28 FAMILY | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET AGGREGATION PLUG FORMATION | 36 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| CEBALLOS TARGETS OF TP53 AND MYC DN | 38 | 31 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGG VS IGA DN | 26 | 16 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 AND CD2 DN | 51 | 32 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX UP | 83 | 66 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS UP | 77 | 57 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ZHAN V1 LATE DIFFERENTIATION GENES DN | 15 | 13 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP | 295 | 155 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-140 | 635 | 641 | m8 | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-216 | 303 | 309 | m8 | hsa-miR-216 | UAAUCUCAGCUGGCAACUGUG |
| miR-24 | 92 | 98 | m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-299-5p | 634 | 640 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-342 | 616 | 622 | 1A | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
| miR-365 | 374 | 380 | m8 | hsa-miR-365 | UAAUGCCCCUAAAAAUCCUUAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.