Gene Page: DAO
Summary ?
| GeneID | 1610 |
| Symbol | DAO |
| Synonyms | DAAO|DAMOX|OXDA |
| Description | D-amino-acid oxidase |
| Reference | MIM:124050|HGNC:HGNC:2671|Ensembl:ENSG00000110887|HPRD:15917|Vega:OTTHUMG00000169360 |
| Gene type | protein-coding |
| Map location | 12q24 |
| Pascal p-value | 0.269 |
| Fetal beta | 0.061 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 3 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 46 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2502827 | chr1 | 176044216 | DAO | 1610 | 0.08 | trans | ||
| rs17572651 | chr1 | 218943612 | DAO | 1610 | 8.48E-6 | trans | ||
| snp_a-1919794 | 0 | DAO | 1610 | 0.04 | trans | |||
| rs16829545 | chr2 | 151977407 | DAO | 1610 | 4.606E-11 | trans | ||
| rs3845734 | chr2 | 171125572 | DAO | 1610 | 0.14 | trans | ||
| rs7584986 | chr2 | 184111432 | DAO | 1610 | 2.161E-13 | trans | ||
| rs9810143 | chr3 | 5060209 | DAO | 1610 | 0.08 | trans | ||
| rs4488887 | chr4 | 177695833 | DAO | 1610 | 0.13 | trans | ||
| rs10491487 | chr5 | 80323367 | DAO | 1610 | 2.491E-5 | trans | ||
| rs1368303 | chr5 | 147672388 | DAO | 1610 | 1.667E-6 | trans | ||
| rs1929769 | chr6 | 40765977 | DAO | 1610 | 0.02 | trans | ||
| rs4897051 | chr6 | 149118925 | DAO | 1610 | 0.04 | trans | ||
| rs3118341 | chr9 | 25185518 | DAO | 1610 | 0.11 | trans | ||
| rs9406868 | chr9 | 25223372 | DAO | 1610 | 0.07 | trans | ||
| rs11139334 | chr9 | 84209393 | DAO | 1610 | 0.13 | trans | ||
| rs17263352 | chr9 | 124811572 | DAO | 1610 | 0.15 | trans | ||
| rs7074445 | chr10 | 88088121 | DAO | 1610 | 0.04 | trans | ||
| rs17410931 | chr10 | 98912175 | DAO | 1610 | 0.13 | trans | ||
| rs754765 | chr10 | 132013664 | DAO | 1610 | 0.06 | trans | ||
| rs11612748 | chr12 | 59007670 | DAO | 1610 | 0.04 | trans | ||
| rs17104720 | chr14 | 77127308 | DAO | 1610 | 0.18 | trans | ||
| rs6574467 | chr14 | 79179744 | DAO | 1610 | 0.01 | trans | ||
| rs10146003 | chr14 | 79191170 | DAO | 1610 | 0.01 | trans | ||
| rs16955618 | chr15 | 29937543 | DAO | 1610 | 3.078E-24 | trans | ||
| snp_a-1830894 | 0 | DAO | 1610 | 0.11 | trans | |||
| rs17681878 | chr18 | 34767223 | DAO | 1610 | 0.1 | trans | ||
| rs17085767 | chr18 | 69839397 | DAO | 1610 | 0.05 | trans | ||
| rs6019060 | chr20 | 46965832 | DAO | 1610 | 0.01 | trans | ||
| rs749043 | chr20 | 46995700 | DAO | 1610 | 0.02 | trans |
Section II. Transcriptome annotation
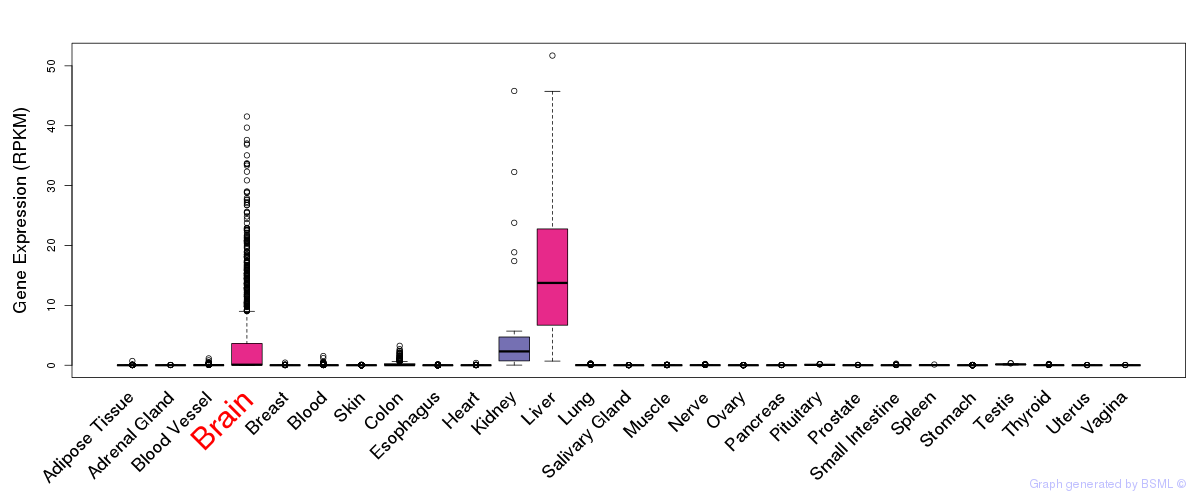
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RHOC | 0.79 | 0.81 |
| MTCP1NB | 0.72 | 0.73 |
| PSME2 | 0.72 | 0.71 |
| FGGY | 0.71 | 0.74 |
| CD63 | 0.71 | 0.73 |
| S100A16 | 0.70 | 0.75 |
| C1orf61 | 0.68 | 0.77 |
| CD58 | 0.67 | 0.67 |
| C1orf54 | 0.67 | 0.73 |
| NKAIN4 | 0.66 | 0.71 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PRDM2 | -0.68 | -0.77 |
| A2BP1 | -0.67 | -0.78 |
| MAGI2 | -0.64 | -0.71 |
| CNNM2 | -0.63 | -0.70 |
| BRPF3 | -0.63 | -0.71 |
| AC016910.1 | -0.63 | -0.73 |
| ANK3 | -0.63 | -0.73 |
| ZBTB38 | -0.63 | -0.71 |
| UBP1 | -0.62 | -0.71 |
| HERC1 | -0.62 | -0.72 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003884 | D-amino-acid oxidase activity | IEA | - | |
| GO:0003884 | D-amino-acid oxidase activity | TAS | 1356107 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004812 | aminoacyl-tRNA ligase activity | IEA | - | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006418 | tRNA aminoacylation for protein translation | IEA | - | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0006551 | leucine metabolic process | IEA | - | |
| GO:0055114 | oxidation reduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005777 | peroxisome | NAS | 2901986 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCINE SERINE AND THREONINE METABOLISM | 31 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG ARGININE AND PROLINE METABOLISM | 54 | 39 | All SZGR 2.0 genes in this pathway |
| KEGG PEROXISOME | 78 | 47 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI UP | 92 | 58 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION DN | 179 | 97 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS POLYSOMY7 UP | 79 | 50 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE DN | 80 | 54 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |