Gene Page: DCTD
Summary ?
| GeneID | 1635 |
| Symbol | DCTD |
| Synonyms | - |
| Description | dCMP deaminase |
| Reference | MIM:607638|HGNC:HGNC:2710|Ensembl:ENSG00000129187|HPRD:09623|Vega:OTTHUMG00000160685 |
| Gene type | protein-coding |
| Map location | 4q35.1 |
| Pascal p-value | 0.406 |
| Sherlock p-value | 0.118 |
| Fetal beta | -0.78 |
| eGene | Anterior cingulate cortex BA24 Cortex Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs3924788 | chr4 | 183807364 | DCTD | 1635 | 0.04 | cis | ||
| rs12507552 | chr4 | 183813424 | DCTD | 1635 | 0.05 | cis | ||
| rs74931353 | 4 | 183854228 | DCTD | ENSG00000129187.10 | 1.21192E-6 | 0.04 | -15139 | gtex_brain_ba24 |
| rs79082694 | 4 | 183854424 | DCTD | ENSG00000129187.10 | 8.98637E-7 | 0.04 | -15335 | gtex_brain_ba24 |
| rs75509458 | 4 | 183857228 | DCTD | ENSG00000129187.10 | 8.98637E-7 | 0.04 | -18139 | gtex_brain_ba24 |
| rs200533947 | 4 | 183804152 | DCTD | ENSG00000129187.10 | 6.39011E-7 | 0.03 | 34937 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
General gene expression (GTEx)
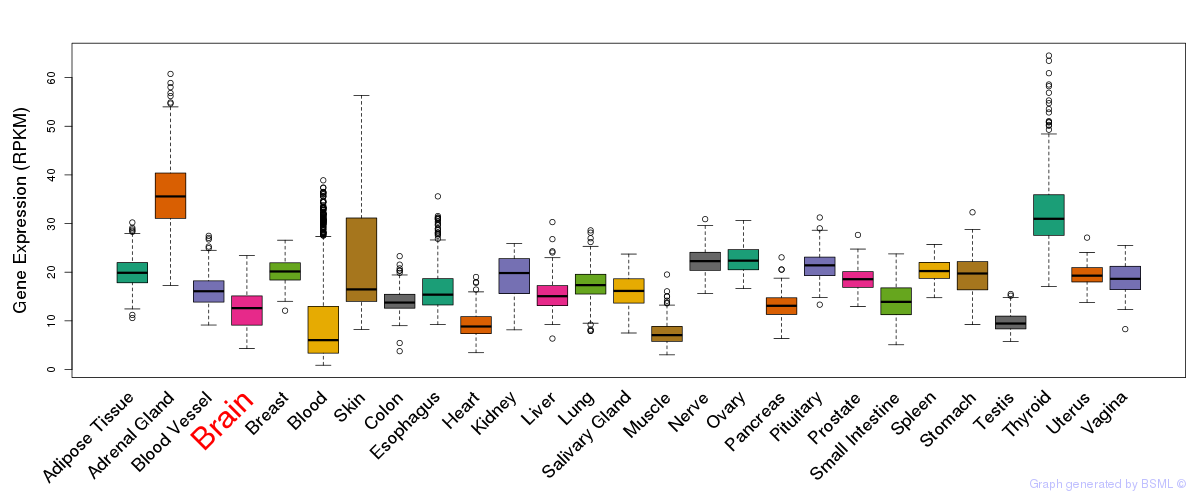
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| COASY | 0.65 | 0.68 |
| FAM82A2 | 0.64 | 0.64 |
| GALNTL1 | 0.62 | 0.66 |
| HS1BP3 | 0.62 | 0.66 |
| FAM120A | 0.62 | 0.66 |
| CASP9 | 0.62 | 0.65 |
| FAM19A5 | 0.61 | 0.65 |
| CLPTM1 | 0.61 | 0.66 |
| TFE3 | 0.61 | 0.67 |
| TRIM9 | 0.60 | 0.68 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM159B | -0.40 | -0.47 |
| AF347015.21 | -0.35 | -0.14 |
| AC087071.1 | -0.34 | -0.21 |
| AC120053.1 | -0.34 | -0.33 |
| AC135724.1 | -0.33 | -0.29 |
| ACSM3 | -0.33 | -0.27 |
| RP9P | -0.33 | -0.36 |
| AC010300.1 | -0.32 | -0.25 |
| AL139819.3 | -0.31 | -0.28 |
| AF347015.18 | -0.31 | -0.13 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PYRIMIDINE METABOLISM | 98 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF NUCLEOTIDES | 72 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME PYRIMIDINE METABOLISM | 24 | 13 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS REPRESSED BY SERUM | 159 | 93 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER EXPRESSION BY COPY NUMBER | 100 | 62 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR UP | 151 | 100 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| LI CYTIDINE ANALOG PATHWAY | 17 | 8 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P6 | 91 | 44 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS | 212 | 121 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS DN | 105 | 63 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO OR GAMMA RADIATION | 15 | 13 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| NUTT GBM VS AO GLIOMA UP | 46 | 34 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ MULTIPLE MYELOMA UP | 35 | 24 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS DN | 72 | 52 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |