Gene Page: DDX3X
Summary ?
| GeneID | 1654 |
| Symbol | DDX3X |
| Synonyms | CAP-Rf|DBX|DDX14|DDX3|HLP2|MRX102 |
| Description | DEAD-box helicase 3, X-linked |
| Reference | MIM:300160|HGNC:HGNC:2745|Ensembl:ENSG00000215301|HPRD:02154|Vega:OTTHUMG00000021369 |
| Gene type | protein-coding |
| Map location | Xp11.3-p11.23 |
| Sherlock p-value | 0.687 |
| Fetal beta | 1.162 |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
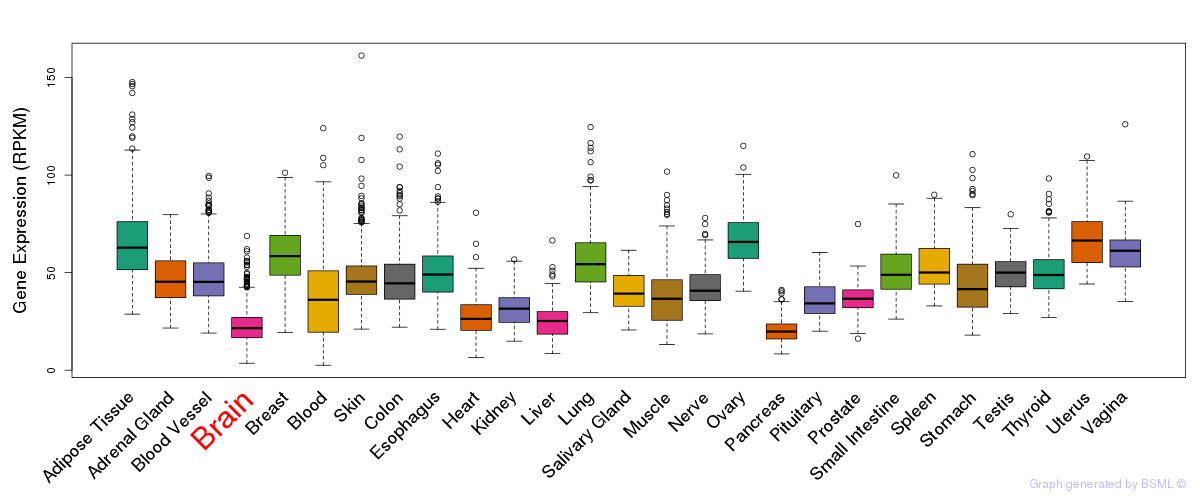
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ACAA2 | 0.86 | 0.84 |
| PRDX6 | 0.82 | 0.75 |
| TNFSF13 | 0.80 | 0.70 |
| GJB6 | 0.80 | 0.71 |
| SP110 | 0.79 | 0.66 |
| AGXT2L1 | 0.78 | 0.73 |
| IL17RB | 0.77 | 0.62 |
| NDRG2 | 0.77 | 0.76 |
| HADHB | 0.77 | 0.78 |
| SAT1 | 0.76 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CACNA1B | -0.64 | -0.72 |
| RTN4RL1 | -0.62 | -0.72 |
| MAPKBP1 | -0.60 | -0.69 |
| BRUNOL5 | -0.60 | -0.70 |
| KIF3C | -0.60 | -0.68 |
| MKL1 | -0.60 | -0.67 |
| HIVEP3 | -0.59 | -0.70 |
| MAST1 | -0.59 | -0.68 |
| RUSC2 | -0.59 | -0.66 |
| NOVA2 | -0.59 | -0.67 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| C20orf24 | PNAS-11 | RIP5 | chromosome 20 open reading frame 24 | Affinity Capture-MS | BioGRID | 17353931 |
| CD40 | Bp50 | CDW40 | MGC9013 | TNFRSF5 | p50 | CD40 molecule, TNF receptor superfamily member 5 | Affinity Capture-MS | BioGRID | 17353931 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | Affinity Capture-MS | BioGRID | 17353931 |
| EIF4A2 | BM-010 | DDX2B | EIF4A | EIF4F | eukaryotic translation initiation factor 4A, isoform 2 | Affinity Capture-MS | BioGRID | 17353931 |
| ELF3 | EPR-1 | ERT | ESE-1 | ESX | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) | Affinity Capture-MS | BioGRID | 17353931 |
| ILK | DKFZp686F1765 | P59 | integrin-linked kinase | Affinity Capture-MS | BioGRID | 17353931 |
| NFKB2 | LYT-10 | LYT10 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | Affinity Capture-MS | BioGRID | 14743216 |
| NUP62 | DKFZp547L134 | FLJ20822 | FLJ43869 | IBSN | MGC841 | SNDI | p62 | nucleoporin 62kDa | DDX3 interacts with Nup62. | BIND | 15507209 |
| PUF60 | FIR | FLJ31379 | RoBPI | SIAHBP1 | poly-U binding splicing factor 60KDa | Affinity Capture-MS | BioGRID | 17353931 |
| SAP18 | 2HOR0202 | MGC27131 | SAP18p | Sin3A-associated protein, 18kDa | Affinity Capture-MS | BioGRID | 17353931 |
| SFRS12 | DKFZp564B176 | MGC133045 | SRrp508 | SRrp86 | splicing factor, arginine/serine-rich 12 | - | HPRD | 14559993 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | Affinity Capture-MS | BioGRID | 17353931 |
| XPO1 | CRM1 | DKFZp686B1823 | exportin 1 (CRM1 homolog, yeast) | DDX3 interacts with CRM1. | BIND | 15507209 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA DN | 136 | 86 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA DN | 146 | 94 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL DN | 101 | 66 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| FALVELLA SMOKERS WITH LUNG CANCER | 80 | 52 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| PETRETTO CARDIAC HYPERTROPHY | 34 | 26 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN | 169 | 102 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D4 | 55 | 37 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR UP | 105 | 73 | All SZGR 2.0 genes in this pathway |
| JI RESPONSE TO FSH DN | 58 | 43 | All SZGR 2.0 genes in this pathway |
| APRELIKOVA BRCA1 TARGETS | 49 | 33 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR UP | 156 | 101 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA CANCER | 83 | 52 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX DN | 54 | 43 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL UP | 193 | 95 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 7 | 76 | 46 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| AZARE NEOPLASTIC TRANSFORMATION BY STAT3 UP | 121 | 70 | All SZGR 2.0 genes in this pathway |
| AZARE STAT3 TARGETS | 24 | 12 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR1 TARGETS UP | 53 | 39 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR133 TARGETS UP | 43 | 27 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |