Gene Page: COCH
Summary ?
| GeneID | 1690 |
| Symbol | COCH |
| Synonyms | COCH-5B2|COCH5B2|DFNA9 |
| Description | cochlin |
| Reference | MIM:603196|HGNC:HGNC:2180|Ensembl:ENSG00000100473|HPRD:04431|Vega:OTTHUMG00000029432 |
| Gene type | protein-coding |
| Map location | 14q11.2-q13 |
| Pascal p-value | 0.837 |
| DEG p-value | DEG:Sanders_2014:DS1_p=-0.148:DS1_beta=0.039400:DS2_p=1.09e-02:DS2_beta=-0.128:DS2_FDR=7.78e-02 |
| eGene | Frontal Cortex BA9 Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Sanders_2013 | Microarray | Whole-genome gene expression profiles using microarrays on lymphoblastoid cell lines (LCLs) from 413 cases and 446 controls. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.047 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs596985 | chr1 | 64277034 | COCH | 1690 | 0 | trans | ||
| rs17131472 | chr1 | 91962966 | COCH | 1690 | 0.07 | trans | ||
| rs6693444 | chr1 | 98690213 | COCH | 1690 | 0 | trans | ||
| rs16838771 | chr1 | 157511227 | COCH | 1690 | 0.02 | trans | ||
| rs3770675 | chr2 | 39480214 | COCH | 1690 | 0.1 | trans | ||
| rs10865148 | chr2 | 39488831 | COCH | 1690 | 0.04 | trans | ||
| rs2302749 | chr2 | 39494196 | COCH | 1690 | 0.04 | trans | ||
| rs10865150 | chr2 | 39529875 | COCH | 1690 | 0.16 | trans | ||
| rs6712274 | chr2 | 39627454 | COCH | 1690 | 0.14 | trans | ||
| rs16823058 | chr2 | 170317903 | COCH | 1690 | 0.02 | trans | ||
| rs12466062 | chr2 | 188137037 | COCH | 1690 | 0.1 | trans | ||
| rs17044102 | chr3 | 17889838 | COCH | 1690 | 0.19 | trans | ||
| rs6793457 | chr3 | 62053678 | COCH | 1690 | 0.02 | trans | ||
| rs10937395 | chr3 | 189336369 | COCH | 1690 | 0.19 | trans | ||
| rs491989 | chr5 | 31452998 | COCH | 1690 | 0.02 | trans | ||
| rs6893045 | chr5 | 31473728 | COCH | 1690 | 0.15 | trans | ||
| rs620498 | chr5 | 133869269 | COCH | 1690 | 0.07 | trans | ||
| rs17101786 | chr5 | 143702446 | COCH | 1690 | 0.04 | trans | ||
| rs10055809 | chr5 | 143766245 | COCH | 1690 | 0.02 | trans | ||
| rs283063 | chr6 | 118621313 | COCH | 1690 | 4.68E-5 | trans | ||
| rs283058 | chr6 | 118625224 | COCH | 1690 | 0.15 | trans | ||
| rs1411442 | chr9 | 72210550 | COCH | 1690 | 0.08 | trans | ||
| rs1411441 | chr9 | 72210608 | COCH | 1690 | 0.07 | trans | ||
| rs17143455 | chr10 | 8368252 | COCH | 1690 | 0 | trans | ||
| rs7081986 | chr10 | 90807242 | COCH | 1690 | 0.04 | trans | ||
| rs7969972 | chr12 | 15763898 | COCH | 1690 | 0.12 | trans | ||
| rs4644683 | chr12 | 15780762 | COCH | 1690 | 0.1 | trans | ||
| rs4764219 | chr12 | 15812187 | COCH | 1690 | 0.01 | trans | ||
| rs7295589 | chr12 | 15816182 | COCH | 1690 | 0.1 | trans | ||
| rs16945668 | chr12 | 115934729 | COCH | 1690 | 0.19 | trans | ||
| rs10047895 | chr14 | 57512171 | COCH | 1690 | 0.15 | trans | ||
| rs1953844 | chr14 | 82453917 | COCH | 1690 | 0.1 | trans | ||
| rs7177212 | chr15 | 34425413 | COCH | 1690 | 0.02 | trans | ||
| rs284907 | chr15 | 76741568 | COCH | 1690 | 0.13 | trans | ||
| rs284898 | chr15 | 76750694 | COCH | 1690 | 0.06 | trans | ||
| rs16959711 | chr16 | 56597605 | COCH | 1690 | 0.1 | trans | ||
| rs8056861 | chr16 | 56921039 | COCH | 1690 | 0.08 | trans | ||
| rs443222 | chr16 | 83889679 | COCH | 1690 | 0.15 | trans | ||
| rs824575 | chr16 | 83891459 | COCH | 1690 | 0.15 | trans | ||
| rs7503018 | chr17 | 37931219 | COCH | 1690 | 0.18 | trans | ||
| rs11873703 | chr18 | 61448702 | COCH | 1690 | 0.03 | trans | ||
| rs1883665 | chrX | 17650382 | COCH | 1690 | 3.018E-8 | trans | ||
| rs7062071 | chrX | 17724830 | COCH | 1690 | 9.796E-5 | trans |
Section II. Transcriptome annotation
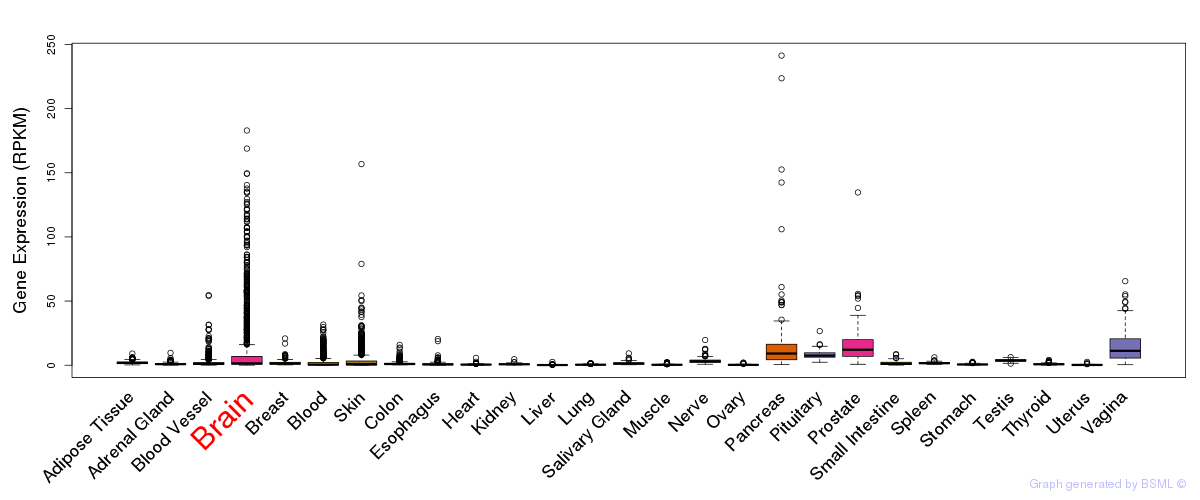
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0007605 | sensory perception of sound | TAS | 9806553 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005578 | proteinaceous extracellular matrix | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| SAMOLS TARGETS OF KHSV MIRNAS UP | 8 | 7 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL DN | 118 | 79 | All SZGR 2.0 genes in this pathway |
| KORKOLA YOLK SAC TUMOR | 62 | 33 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER B | 48 | 22 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION UP | 140 | 83 | All SZGR 2.0 genes in this pathway |
| ABE INNER EAR | 48 | 26 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| YEGNASUBRAMANIAN PROSTATE CANCER | 128 | 60 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| VICENT METASTASIS UP | 14 | 11 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 DN | 228 | 114 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME3 AND H3K27ME3 | 142 | 103 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G23 UP | 52 | 35 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K4ME3 AND H3K27ME3 | 210 | 148 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 3 UP | 170 | 97 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 4 UP | 112 | 64 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 UP | 140 | 81 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 7 UP | 118 | 68 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP2 | 60 | 38 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP C | 92 | 60 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS DN | 148 | 88 | All SZGR 2.0 genes in this pathway |
| NABA ECM GLYCOPROTEINS | 196 | 99 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-137 | 797 | 804 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-378 | 234 | 240 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.