Gene Page: DLD
Summary ?
| GeneID | 1738 |
| Symbol | DLD |
| Synonyms | DLDD|DLDH|E3|GCSL|LAD|PHE3 |
| Description | dihydrolipoamide dehydrogenase |
| Reference | MIM:238331|HGNC:HGNC:2898|Ensembl:ENSG00000091140|HPRD:02006|Vega:OTTHUMG00000154813 |
| Gene type | protein-coding |
| Map location | 7q31-q32 |
| Pascal p-value | 0.961 |
| Sherlock p-value | 0.074 |
| Fetal beta | -0.61 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cortex Frontal Cortex BA9 |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_mitochondria G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:PheWAS | Phenome-wide association studies (PheWAS) | 157 SNPs associated with schizophrenia | 0 |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias,schizotypal | Click to show details |
Section I. Genetics and epigenetics annotation
 CV:PheWAS
CV:PheWAS
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05663655 | 7 | 107532014 | DLD | 3.56E-8 | -0.011 | 1.04E-5 | DMG:Jaffe_2016 |
| cg18679886 | 7 | 107531975 | DLD | 5.23E-8 | -0.009 | 1.37E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
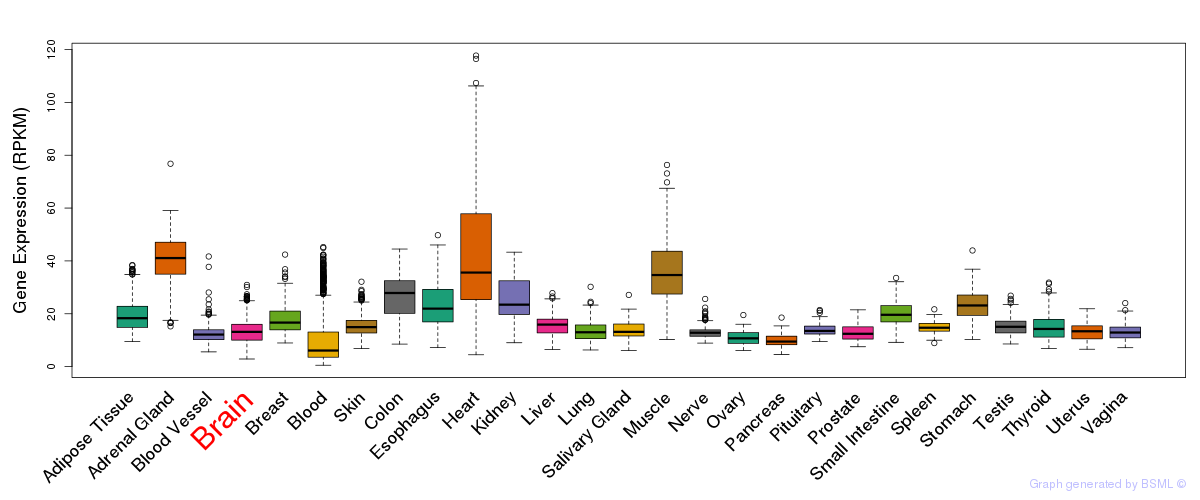
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SIX3 | 0.96 | 0.31 |
| C10orf41 | 0.93 | 0.37 |
| PBX3 | 0.90 | 0.54 |
| ZNF503 | 0.89 | 0.52 |
| NTN1 | 0.88 | 0.39 |
| RXRG | 0.88 | 0.32 |
| AC010997.1 | 0.88 | 0.52 |
| POU3F4 | 0.88 | 0.45 |
| AP000911.1 | 0.87 | 0.34 |
| SP9 | 0.86 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CHPT1 | -0.27 | -0.22 |
| CDADC1 | -0.24 | 0.16 |
| AF347015.31 | -0.22 | -0.44 |
| C5orf53 | -0.22 | -0.29 |
| COPZ2 | -0.22 | -0.43 |
| MT-CO2 | -0.22 | -0.45 |
| PTH1R | -0.22 | -0.23 |
| MYL3 | -0.21 | -0.41 |
| C16orf89 | -0.21 | -0.36 |
| COX4I2 | -0.21 | -0.40 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOLYSIS GLUCONEOGENESIS | 62 | 41 | All SZGR 2.0 genes in this pathway |
| KEGG CITRATE CYCLE TCA CYCLE | 32 | 23 | All SZGR 2.0 genes in this pathway |
| KEGG GLYCINE SERINE AND THREONINE METABOLISM | 31 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG VALINE LEUCINE AND ISOLEUCINE DEGRADATION | 44 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG PYRUVATE METABOLISM | 40 | 26 | All SZGR 2.0 genes in this pathway |
| ST JNK MAPK PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | 48 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME TCA CYCLE AND RESPIRATORY ELECTRON TRANSPORT | 141 | 85 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | 13 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME CITRIC ACID CYCLE TCA CYCLE | 26 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | 17 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME PYRUVATE METABOLISM | 19 | 14 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| RAHMAN TP53 TARGETS PHOSPHORYLATED | 21 | 18 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6 | 189 | 112 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| WONG MITOCHONDRIA GENE MODULE | 217 | 122 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |