Gene Page: ELAVL1
Summary ?
| GeneID | 1994 |
| Symbol | ELAVL1 |
| Synonyms | ELAV1|HUR|Hua|MelG |
| Description | ELAV like RNA binding protein 1 |
| Reference | MIM:603466|HGNC:HGNC:3312|Ensembl:ENSG00000066044|HPRD:16025|Vega:OTTHUMG00000182470 |
| Gene type | protein-coding |
| Map location | 19p13.2 |
| Pascal p-value | 0.146 |
| Sherlock p-value | 0.008 |
| Fetal beta | 0.831 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
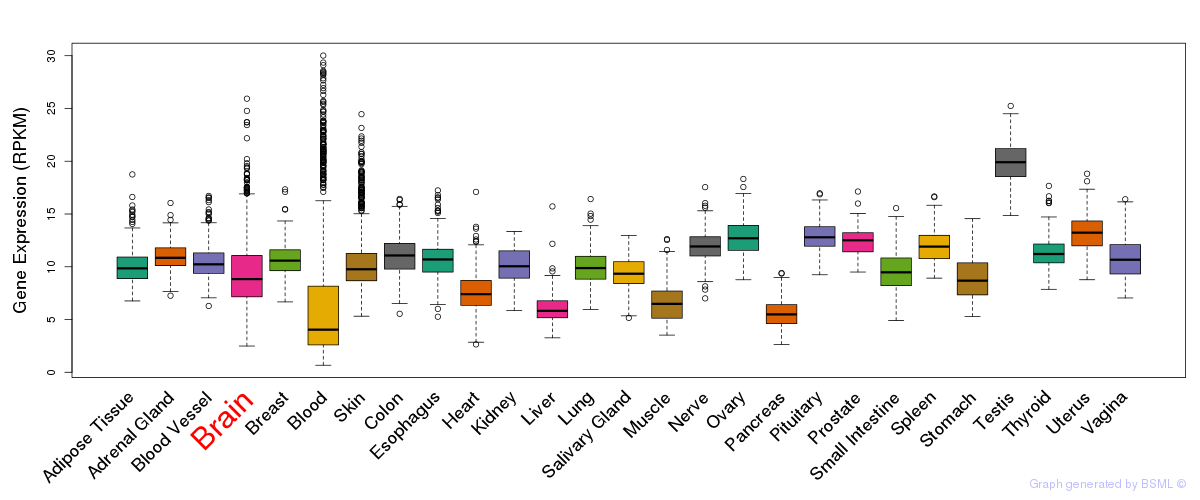
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DAB1 | 0.96 | 0.87 |
| AFF3 | 0.95 | 0.94 |
| TIAM2 | 0.95 | 0.91 |
| BACH2 | 0.94 | 0.92 |
| SEZ6 | 0.94 | 0.94 |
| SSBP2 | 0.94 | 0.89 |
| DACT1 | 0.93 | 0.90 |
| FAT4 | 0.93 | 0.90 |
| LRCH1 | 0.93 | 0.92 |
| ANKRD44 | 0.93 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SERPINB6 | -0.65 | -0.73 |
| C5orf53 | -0.62 | -0.76 |
| AIFM3 | -0.62 | -0.80 |
| HSD17B14 | -0.61 | -0.82 |
| HEPN1 | -0.61 | -0.74 |
| TNFSF12 | -0.61 | -0.66 |
| AC007405.8 | -0.61 | -0.66 |
| SLC16A11 | -0.61 | -0.67 |
| LHPP | -0.60 | -0.59 |
| RAMP1 | -0.60 | -0.80 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANP32A | C15orf1 | I1PP2A | LANP | MAPM | MGC119787 | MGC150373 | PHAP1 | PHAPI | PP32 | acidic (leucine-rich) nuclear phosphoprotein 32 family, member A | - | HPRD | 11018049 |11565755 |11729309 |
| ANP32B | APRIL | PHAPI2 | SSP29 | acidic (leucine-rich) nuclear phosphoprotein 32 family, member B | - | HPRD | 11018049 |11565755 |11729309 |
| CARM1 | PRMT4 | coactivator-associated arginine methyltransferase 1 | - | HPRD | 12237300 |
| HNRNPA1 | HNRPA1 | MGC102835 | heterogeneous nuclear ribonucleoprotein A1 | - | HPRD | 11565755 |
| HNRNPAB | ABBP1 | FLJ40338 | HNRPAB | heterogeneous nuclear ribonucleoprotein A/B | - | HPRD | 11565755 |
| KPNA1 | IPOA5 | NPI-1 | RCH2 | SRP1 | karyopherin alpha 1 (importin alpha 5) | - | HPRD | 15342649 |
| KPNB1 | IMB1 | IPOB | Impnb | MGC2155 | MGC2156 | MGC2157 | NTF97 | karyopherin (importin) beta 1 | - | HPRD | 15037768 |15342649 |
| PAIP2 | MGC72018 | PAIP2A | poly(A) binding protein interacting protein 2 | - | HPRD | 15175342 |
| PTMA | MGC104802 | TMSA | prothymosin, alpha | ELAVL1 (HuR) interacts with PMTA (ProT-alpha) mRNA. | BIND | 15861128 |
| SET | 2PP2A | I2PP2A | IGAAD | IPP2A2 | PHAPII | TAF-I | TAF-IBETA | SET nuclear oncogene | - | HPRD | 11018049 |11565755 |
| SFRS12 | DKFZp564B176 | MGC133045 | SRrp508 | SRrp86 | splicing factor, arginine/serine-rich 12 | - | HPRD | 14559993 |
| TNPO1 | IPO2 | KPNB2 | MIP | MIP1 | TRN | transportin 1 | - | HPRD | 15037768 |15342649 |
| TNPO2 | FLJ12155 | IPO3 | KPNB2B | TRN2 | transportin 2 (importin 3, karyopherin beta 2b) | - | HPRD | 11729309 |14981248 |15037768 |
| XPO1 | CRM1 | DKFZp686B1823 | exportin 1 (CRM1 homolog, yeast) | - | HPRD,BioGRID | 11565755 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA VEGF PATHWAY | 29 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | 84 | 50 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 2 DN | 77 | 46 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER UP | 142 | 96 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW UP | 162 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH UP | 147 | 101 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA UP | 146 | 104 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER SOX9 TARGETS IN PROSTATE DEVELOPMENT DN | 45 | 33 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| PETRETTO HEART MASS QTL CIS DN | 24 | 15 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 24HR | 43 | 30 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS DN | 314 | 188 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MOLENAAR TARGETS OF CCND1 AND CDK4 DN | 58 | 25 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS POLYSOMY7 UP | 79 | 50 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP | 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| KAMMINGA SENESCENCE | 41 | 26 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR133 TARGETS UP | 43 | 27 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |