Gene Page: EPOR
Summary ?
| GeneID | 2057 |
| Symbol | EPOR |
| Synonyms | EPO-R |
| Description | erythropoietin receptor |
| Reference | MIM:133171|HGNC:HGNC:3416|Ensembl:ENSG00000187266|HPRD:00587|Vega:OTTHUMG00000182027 |
| Gene type | protein-coding |
| Map location | 19p13.2 |
| Pascal p-value | 0.8 |
| Fetal beta | -0.488 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
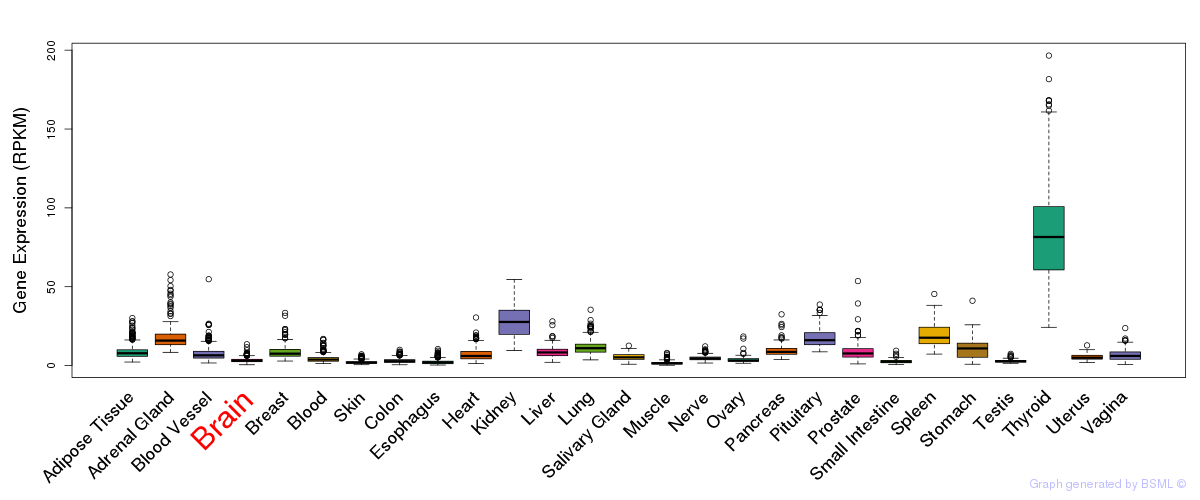
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF148 | 0.94 | 0.96 |
| IREB2 | 0.94 | 0.95 |
| GSPT1 | 0.94 | 0.94 |
| GTF3C4 | 0.94 | 0.95 |
| EPM2AIP1 | 0.94 | 0.95 |
| PPP2R5E | 0.94 | 0.94 |
| BCLAF1 | 0.94 | 0.95 |
| ZMYM2 | 0.93 | 0.94 |
| ZMYM4 | 0.93 | 0.94 |
| DIS3 | 0.93 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.74 | -0.85 |
| AF347015.31 | -0.73 | -0.83 |
| FXYD1 | -0.71 | -0.82 |
| MT-CYB | -0.71 | -0.81 |
| AF347015.33 | -0.71 | -0.79 |
| HIGD1B | -0.70 | -0.82 |
| AF347015.8 | -0.70 | -0.82 |
| IFI27 | -0.69 | -0.81 |
| AF347015.27 | -0.69 | -0.79 |
| AIFM3 | -0.69 | -0.73 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005085 | guanyl-nucleotide exchange factor activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 11781573 |15644415 | |
| GO:0004900 | erythropoietin receptor activity | IDA | 2163696 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | IEA | Brain (GO term level: 7) | - |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0007165 | signal transduction | NAS | 1668606 | |
| GO:0007507 | heart development | IEA | - | |
| GO:0007264 | small GTPase mediated signal transduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 1848667 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATXN2L | A2D | A2LG | A2LP | A2RP | ataxin 2-like | - | HPRD,BioGRID | 11784712 |
| CISH | CIS | CIS-1 | G18 | SOCS | cytokine inducible SH2-containing protein | - | HPRD | 9774439 |
| CISH | CIS | CIS-1 | G18 | SOCS | cytokine inducible SH2-containing protein | - | HPRD,BioGRID | 7796808 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | - | HPRD,BioGRID | 11443118 |
| CSF2RB | CD131 | CDw131 | IL3RB | IL5RB | colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | - | HPRD | 12488507 |
| EPO | EP | MGC138142 | erythropoietin | - | HPRD,BioGRID | 9808045 |10318834 |
| EPO | EP | MGC138142 | erythropoietin | Interaction between EPOR (PDB ID: 1EER_C) and EPO (PDB ID: 1EER_A). | BIND | 9774108 |
| EPO | EP | MGC138142 | erythropoietin | Interaction between EPOR (PDB ID: 1EER_B) and EPO (PDB ID: 1EER_A). | BIND | 9774108 |
| EPOR | MGC138358 | erythropoietin receptor | Interaction between EPOR (PDB ID: 1ERN_B) and EPOR (PDB ID: 1ERN_A). | BIND | 8662530 |9974392 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | - | HPRD | 10455108 |
| GNAI1 | Gi | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | Reconstituted Complex | BioGRID | 12538595 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | EPOR interacts with RACK-1. This interaction was modelled on a demonstrated interaction between EPOR from an unspecified species and human RACK-1. | BIND | 12960323 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | - | HPRD,BioGRID | 12960323 |
| GRAP | MGC64880 | GRB2-related adaptor protein | - | HPRD,BioGRID | 8647802 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 7534299 |
| IRS2 | - | insulin receptor substrate 2 | - | HPRD,BioGRID | 9334184 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | - | HPRD,BioGRID | 8343951 |11779507 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | - | HPRD | 11124255 |
| KITLG | DKFZp686F2250 | KL-1 | Kitl | MGF | SCF | SF | SHEP7 | KIT ligand | - | HPRD | 11124255 |
| KLK3 | APS | KLK2A1 | PSA | hK3 | kallikrein-related peptidase 3 | - | HPRD | 10374881 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | - | HPRD,BioGRID | 9573010 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | - | HPRD | 12538595 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | - | HPRD | 12538595 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD,BioGRID | 7559499 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | - | HPRD,BioGRID | 8639815 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | - | HPRD,BioGRID | 7889566 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 8400282 |
| SOCS2 | CIS2 | Cish2 | SOCS-2 | SSI-2 | SSI2 | STATI2 | suppressor of cytokine signaling 2 | - | HPRD,BioGRID | 11781573 |
| SOCS3 | ATOD4 | CIS3 | Cish3 | MGC71791 | SOCS-3 | SSI-3 | SSI3 | suppressor of cytokine signaling 3 | - | HPRD,BioGRID | 10882725 |12027890 |
| STAT5A | MGF | STAT5 | signal transducer and activator of transcription 5A | - | HPRD,BioGRID | 8977232 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | - | HPRD,BioGRID | 9852052 |
| SYP | - | synaptophysin | Reconstituted Complex | BioGRID | 7534299 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD,BioGRID | 9162069 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| KEGG HEMATOPOIETIC CELL LINEAGE | 88 | 60 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EPO PATHWAY | 19 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EPONFKB PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| PID KIT PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID EPO PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR UP | 162 | 116 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER PAX8 PPARG UP | 44 | 29 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD UP | 64 | 39 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER CLUSTER 1 | 51 | 33 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| LIU NASOPHARYNGEAL CARCINOMA | 70 | 38 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 120 MCF10A | 43 | 28 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 20 MCF10A | 21 | 10 | All SZGR 2.0 genes in this pathway |
| BASSO B LYMPHOCYTE NETWORK | 143 | 96 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO ESTRADIOL DN | 82 | 52 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| MUNSHI MULTIPLE MYELOMA UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS MATURE CELL | 293 | 160 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS UP | 85 | 50 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A5 | 70 | 32 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 15 | 35 | 23 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS EARLY DN | 38 | 27 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS LATE DN | 38 | 25 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS DN | 116 | 74 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN VINCRISTINE RESISTANCE ALL UP | 27 | 16 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA NS UP | 162 | 103 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 2HR DN | 55 | 35 | All SZGR 2.0 genes in this pathway |