Gene Page: AKT1
Summary ?
| GeneID | 207 |
| Symbol | AKT1 |
| Synonyms | AKT|CWS6|PKB|PKB-ALPHA|PRKBA|RAC|RAC-ALPHA |
| Description | v-akt murine thymoma viral oncogene homolog 1 |
| Reference | MIM:164730|HGNC:HGNC:391|Ensembl:ENSG00000142208|HPRD:01261|Vega:OTTHUMG00000170795 |
| Gene type | protein-coding |
| Map location | 14q32.32 |
| Pascal p-value | 0.423 |
| Sherlock p-value | 0.761 |
| Fetal beta | 1.011 |
| DMG | 2 (# studies) |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 31.3585 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AKT1 | chr14 | 105239278 | C | T | NM_001014431 NM_001014432 NM_005163 | p.370R>H p.370R>H p.370R>H | missense missense missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04971812 | 14 | 105236654 | AKT1 | 1.23E-4 | 0.471 | 0.03 | DMG:Wockner_2014 |
| cg01749142 | 14 | 105262305 | AKT1 | 1.715E-4 | -0.319 | 0.033 | DMG:Wockner_2014 |
| cg03734874 | 14 | 105071382 | AKT1 | 4.993E-4 | -4.762 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
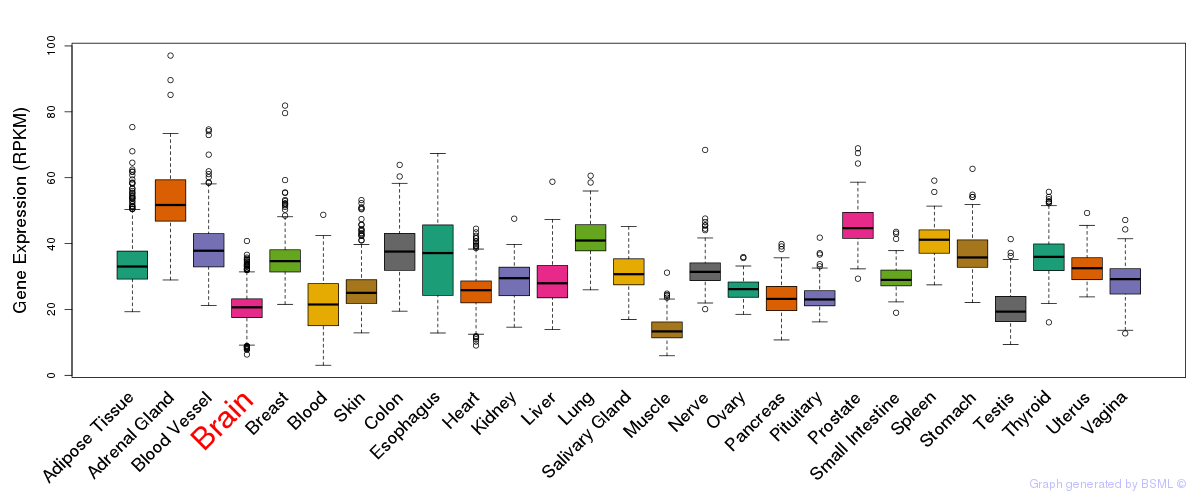
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MAN2A2 | 0.86 | 0.84 |
| IPO13 | 0.86 | 0.82 |
| ABCA2 | 0.85 | 0.83 |
| SLC6A8 | 0.83 | 0.84 |
| FAM102A | 0.82 | 0.77 |
| SEMA4D | 0.82 | 0.81 |
| ANGEL1 | 0.82 | 0.81 |
| NIPAL3 | 0.81 | 0.81 |
| KLHL21 | 0.81 | 0.80 |
| TMEM189 | 0.80 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SNRPG | -0.49 | -0.63 |
| C9orf46 | -0.49 | -0.55 |
| FAM36A | -0.48 | -0.50 |
| RPL31 | -0.46 | -0.55 |
| RPL13AP22 | -0.45 | -0.67 |
| SPINK8 | -0.45 | -0.52 |
| RPL35 | -0.45 | -0.55 |
| RPL24 | -0.45 | -0.53 |
| RPL35A | -0.45 | -0.57 |
| TBCA | -0.44 | -0.46 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0030235 | nitric-oxide synthase regulator activity | IMP | 10376603 | |
| GO:0005351 | sugar:hydrogen symporter activity | IEA | - | |
| GO:0005524 | ATP binding | IC | 11994271 | |
| GO:0004674 | protein serine/threonine kinase activity | EXP | 9829964 |10490848 |11274386 | |
| GO:0004674 | protein serine/threonine kinase activity | IDA | 11994271 |16139227 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0042802 | identical protein binding | IPI | 7891724 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000060 | protein import into nucleus, translocation | IMP | 16280327 | |
| GO:0001568 | blood vessel development | IEA | - | |
| GO:0001890 | placenta development | IEA | - | |
| GO:0006417 | regulation of translation | IEA | - | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | TAS | 10570282 | |
| GO:0006464 | protein modification process | TAS | 10570282 | |
| GO:0007242 | intracellular signaling cascade | IDA | 14749367 | |
| GO:0007281 | germ cell development | IEA | - | |
| GO:0006006 | glucose metabolic process | IEA | - | |
| GO:0005978 | glycogen biosynthetic process | IEA | - | |
| GO:0005975 | carbohydrate metabolic process | IEA | - | |
| GO:0008286 | insulin receptor signaling pathway | IMP | 8978681 | |
| GO:0008633 | activation of pro-apoptotic gene products | EXP | 15231831 | |
| GO:0008637 | apoptotic mitochondrial changes | IEA | - | |
| GO:0008643 | carbohydrate transport | IEA | - | |
| GO:0009408 | response to heat | TAS | 10958679 | |
| GO:0009725 | response to hormone stimulus | IEA | - | |
| GO:0015758 | glucose transport | IEA | - | |
| GO:0048009 | insulin-like growth factor receptor signaling pathway | IMP | 8978681 | |
| GO:0006954 | inflammatory response | IEA | - | |
| GO:0006924 | activated T cell apoptosis | IMP | 14749367 | |
| GO:0016567 | protein ubiquitination | IEA | - | |
| GO:0018105 | peptidyl-serine phosphorylation | IDA | 16139227 | |
| GO:0051000 | positive regulation of nitric-oxide synthase activity | IMP | 10376603 | |
| GO:0040018 | positive regulation of multicellular organism growth | IEA | - | |
| GO:0034405 | response to fluid shear stress | IMP | 10376603 | |
| GO:0030163 | protein catabolic process | IEA | - | |
| GO:0043491 | protein kinase B signaling cascade | IEA | - | |
| GO:0042640 | anagen | IEA | - | |
| GO:0045884 | regulation of survival gene product expression | IEA | - | |
| GO:0046777 | protein amino acid autophosphorylation | TAS | 16280327 | |
| GO:0045429 | positive regulation of nitric oxide biosynthetic process | IMP | 10376603 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005819 | spindle | IEA | - | |
| GO:0005829 | cytosol | EXP | 9381178 |9812896 |10485710 |10698680 |11715018 |12167717 |12244303 |12524439 |15183529 |15718470 | |
| GO:0005856 | cytoskeleton | IDA | 18029348 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | EXP | 9829964 |10490848 |11274386 | |
| GO:0005737 | cytoplasm | ISS | - | |
| GO:0030027 | lamellipodium | IEA | - | |
| GO:0005886 | plasma membrane | EXP | 10698680 |15808505 | |
| GO:0005886 | plasma membrane | IDA | 14749367 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKTIP | FT1 | FTS | AKT interacting protein | - | HPRD,BioGRID | 14749367 |
| APPL1 | APPL | DIP13alpha | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 | APPL interacts with Akt1. | BIND | 10490823 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Akt phosphorylates AR. | BIND | 11404460 |12621049 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Affinity Capture-Western Reconstituted Complex | BioGRID | 11404460 |
| ARFIP2 | POR1 | ADP-ribosylation factor interacting protein 2 | Two-hybrid | BioGRID | 9312003 |
| ARHGAP29 | PARG1 | RP11-255E17.1 | Rho GTPase activating protein 29 | Two-hybrid | BioGRID | 14749388 |
| BAD | BBC2 | BCL2L8 | BCL2-associated agonist of cell death | Reconstituted Complex | BioGRID | 11500364 |
| BRAF | B-RAF1 | BRAF1 | FLJ95109 | MGC126806 | MGC138284 | RAFB1 | v-raf murine sarcoma viral oncogene homolog B1 | - | HPRD,BioGRID | 10869359 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Reconstituted Complex | BioGRID | 10542266 |
| CARHSP1 | CRHSP-24 | CSDC1 | MGC111446 | calcium regulated heat stable protein 1, 24kDa | Reconstituted Complex | BioGRID | 15910284 |
| CDKN1B | CDKN4 | KIP1 | MEN1B | MEN4 | P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | Affinity Capture-Western Biochemical Activity | BioGRID | 12042314 |
| CHN2 | ARHGAP3 | BCH | MGC138360 | RHOGAP3 | chimerin (chimaerin) 2 | Reconstituted Complex | BioGRID | 11278894 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | - | HPRD,BioGRID | 10485710 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | CK2 interacts with and phosphorylates Akt. This interaction was modeled on a demonstrated interaction between human CK2 and Akt from an unspecified species and mouse. | BIND | 15818404 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | Two-hybrid | BioGRID | 14499622 |
| FOXO4 | AFX | AFX1 | MGC120490 | MLLT7 | forkhead box O4 | - | HPRD | 11313479 |
| FOXO4 | AFX | AFX1 | MGC120490 | MLLT7 | forkhead box O4 | AKT1 interacts with MLLT7 (FOXO4). This interaction was modelled on a demonstrated interaction between human AKT1 and FOXO4 from an unspecified species. | BIND | 15688030 |
| FRAP1 | FLJ44809 | FRAP | FRAP2 | MTOR | RAFT1 | RAPT1 | FK506 binding protein 12-rapamycin associated protein 1 | Protein-peptide Reconstituted Complex | BioGRID | 10910062 |14970221 |15718470 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | - | HPRD,BioGRID | 11782427 |
| GRB10 | GRB-IR | Grb-10 | IRBP | KIAA0207 | MEG1 | RSS | growth factor receptor-bound protein 10 | - | HPRD | 11809791 |
| GSK3B | - | glycogen synthase kinase 3 beta | AKT1 interacts with and phosphorylates GSK3B. This interaction was modelled on a demonstrated interaction between human AKT1 and GSK3B from an unspecified species. | BIND | 15688030 |
| GSK3B | - | glycogen synthase kinase 3 beta | Reconstituted Complex | BioGRID | 16009706 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD,BioGRID | 10995457 |
| HSP90AB1 | D6S182 | FLJ26984 | HSP90-BETA | HSP90B | HSPC2 | HSPCB | heat shock protein 90kDa alpha (cytosolic), class B member 1 | Affinity Capture-Western | BioGRID | 10995457 |
| HSPB1 | CMT2F | DKFZp586P1322 | HMN2B | HS.76067 | HSP27 | HSP28 | Hsp25 | SRP27 | heat shock 27kDa protein 1 | Affinity Capture-Western | BioGRID | 11042204 |
| HTT | HD | IT15 | huntingtin | Huntingtin is serine-phosphorylated by Akt. | BIND | 12062094 |
| ILK | DKFZp686F1765 | P59 | integrin-linked kinase | - | HPRD,BioGRID | 9736715 |
| IMPDH2 | IMPD2 | IMPDH-II | IMP (inosine monophosphate) dehydrogenase 2 | The pleckstrin homology domain of PKB/Akt-alpha interacts with IMPDH type II. | BIND | 10930578 |
| IMPDH2 | IMPD2 | IMPDH-II | IMP (inosine monophosphate) dehydrogenase 2 | - | HPRD,BioGRID | 10930578 |
| IRAK1 | IRAK | pelle | interleukin-1 receptor-associated kinase 1 | - | HPRD,BioGRID | 11976320 |
| IRS1 | HIRS-1 | insulin receptor substrate 1 | - | HPRD,BioGRID | 10497255 |
| KRT10 | CK10 | K10 | KPP | keratin 10 | - | HPRD,BioGRID | 11585925 |
| KTN1 | CG1 | KIAA0004 | KNT | MGC133337 | MU-RMS-40.19 | kinectin 1 (kinesin receptor) | Two-hybrid | BioGRID | 11162552 |
| MAP2K4 | JNKK | JNKK1 | MAPKK4 | MEK4 | MKK4 | PRKMK4 | SEK1 | SERK1 | mitogen-activated protein kinase kinase 4 | - | HPRD,BioGRID | 11707464 |
| MAP3K11 | MGC17114 | MLK-3 | MLK3 | PTK1 | SPRK | mitogen-activated protein kinase kinase kinase 11 | - | HPRD,BioGRID | 12458207 |
| MAP3K8 | COT | EST | ESTF | FLJ10486 | TPL2 | Tpl-2 | c-COT | mitogen-activated protein kinase kinase kinase 8 | - | HPRD,BioGRID | 12138205 |
| MAPK14 | CSBP1 | CSBP2 | CSPB1 | EXIP | Mxi2 | PRKM14 | PRKM15 | RK | SAPK2A | p38 | p38ALPHA | mitogen-activated protein kinase 14 | Affinity Capture-Western Biochemical Activity | BioGRID | 11042204 |
| MAPK8IP1 | IB1 | JIP-1 | JIP1 | PRKM8IP | mitogen-activated protein kinase 8 interacting protein 1 | - | HPRD,BioGRID | 12194869 |
| MAPKAP1 | JC310 | MGC2745 | MIP1 | SIN1 | SIN1b | SIN1g | mitogen-activated protein kinase associated protein 1 | Affinity Capture-Western | BioGRID | 16962653 |
| MAPKAPK2 | MK2 | mitogen-activated protein kinase-activated protein kinase 2 | - | HPRD,BioGRID | 11042204 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | HDM2 interacts with and is phosphorylated by Akt. | BIND | 11960368 |
| METTL1 | C12orf1 | FLJ95748 | TRM8 | YDL201w | methyltransferase like 1 | Mettl1 interacts with and is phosphorylated by PKB-alpha (Akt) | BIND | 15861136 |
| MTCP1 | C6.1B | mature T-cell proliferation 1 | - | HPRD,BioGRID | 11707444 |
| NCF1C | SH3PXD1C | neutrophil cytosolic factor 1C pseudogene | Akt interaction with and phosphorylation of p47-phox | BIND | 12734380 |
| NR4A1 | GFRP1 | HMR | MGC9485 | N10 | NAK-1 | NGFIB | NP10 | NUR77 | TR3 | nuclear receptor subfamily 4, group A, member 1 | - | HPRD,BioGRID | 11274386 |
| PAK1 | MGC130000 | MGC130001 | PAKalpha | p21 protein (Cdc42/Rac)-activated kinase 1 | Reconstituted Complex | BioGRID | 14749719 |
| PAK6 | PAK5 | p21 protein (Cdc42/Rac)-activated kinase 6 | Reconstituted Complex | BioGRID | 11773441 |
| PDK1 | - | pyruvate dehydrogenase kinase, isozyme 1 | PDK1 interacts with and phosphorylates Akt1. This interaction was modeled on a demonstrated interaction between PDK1 and Akt1, both from unspecified species. | BIND | 15678105 |
| PDPK1 | MGC20087 | MGC35290 | PDK1 | PRO0461 | 3-phosphoinositide dependent protein kinase-1 | - | HPRD,BioGRID | 11825911 |
| PDPK1 | MGC20087 | MGC35290 | PDK1 | PRO0461 | 3-phosphoinositide dependent protein kinase-1 | PDK1 phosphorylates Akt1. | BIND | 15718470 |
| PEA15 | HMAT1 | HUMMAT1H | MAT1 | MAT1H | PEA-15 | PED | phosphoprotein enriched in astrocytes 15 | Akt interacts with PED. | BIND | 12808093 |
| PHLPP | MGC161555 | PHLPP1 | PLEKHE1 | SCOP | PH domain and leucine rich repeat protein phosphatase | PHLPP interacts with Akt. This interaction was modelled on a demonstrated interaction between human PHLPP and Akt from an unspecified species. | BIND | 15808505 |
| PIAS2 | MGC102682 | MIZ1 | PIASX | PIASX-ALPHA | PIASX-BETA | SIZ2 | ZMIZ4 | miz | protein inhibitor of activated STAT, 2 | Miz1 interacts with Akt. | BIND | 15580267 |
| PKN2 | MGC150606 | MGC71074 | PAK2 | PRK2 | PRKCL2 | PRO2042 | Pak-2 | protein kinase N2 | - | HPRD,BioGRID | 10926925 |
| PLXNA1 | NOV | NOVP | PLEXIN-A1 | PLXN1 | plexin A1 | - | HPRD,BioGRID | 15187088 |
| PRKCQ | MGC126514 | MGC141919 | PRKCT | nPKC-theta | protein kinase C, theta | - | HPRD,BioGRID | 11410591 |
| PRKCZ | PKC-ZETA | PKC2 | protein kinase C, zeta | - | HPRD,BioGRID | 10085094 |
| PRKDC | DNA-PKcs | DNAPK | DNPK1 | HYRC | HYRC1 | XRCC7 | p350 | protein kinase, DNA-activated, catalytic polypeptide | DNA-PKcs interacts with Akt1. | BIND | 15678105 |
| PTPN1 | PTP1B | protein tyrosine phosphatase, non-receptor type 1 | - | HPRD | 11579209 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD,BioGRID | 10576742 |
| RICS | GC-GAP | GRIT | KIAA0712 | MGC1892 | p200RhoGAP | p250GAP | Rho GTPase-activating protein | Biochemical Activity | BioGRID | 12446789 |
| RICTOR | DKFZp686B11164 | KIAA1999 | MGC39830 | mAVO3 | rapamycin-insensitive companion of mTOR | Reconstituted Complex | BioGRID | 15718470 |
| RPS6KB1 | PS6K | S6K | S6K1 | STK14A | p70(S6K)-alpha | p70-S6K | p70-alpha | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | - | HPRD | 11438723 |
| RPS6KB1 | PS6K | S6K | S6K1 | STK14A | p70(S6K)-alpha | p70-S6K | p70-alpha | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | Co-purification | BioGRID | 15149849 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | Affinity Capture-Western | BioGRID | 16362038 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | Affinity Capture-Western | BioGRID | 16362038 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | Affinity Capture-Western | BioGRID | 16362038 |
| SMAD7 | CRCS3 | FLJ16482 | MADH7 | MADH8 | SMAD family member 7 | Affinity Capture-Western | BioGRID | 16362038 |
| TCL1A | TCL1 | T-cell leukemia/lymphoma 1A | - | HPRD | 10983986 |12009899 |
| TCL1A | TCL1 | T-cell leukemia/lymphoma 1A | Affinity Capture-Western Reconstituted Complex | BioGRID | 10983986 |11707444 |12009899 |
| TCL1A | TCL1 | T-cell leukemia/lymphoma 1A | TCL1 interacts with AKT1. | BIND | 11707444 |
| TCL1B | TML1 | T-cell leukemia/lymphoma 1B | - | HPRD,BioGRID | 10983986 |
| TCL1B | TML1 | T-cell leukemia/lymphoma 1B | TCL1B interacts with AKT1. | BIND | 11707444 |
| TERT | EST2 | TCS1 | TP2 | TRT | hEST2 | telomerase reverse transcriptase | Affinity Capture-Western | BioGRID | 15843522 |
| THEM4 | CTMP | MGC29636 | thioesterase superfamily member 4 | - | HPRD,BioGRID | 11598301 |
| TNFSF11 | CD254 | ODF | OPGL | OPTB2 | RANKL | TRANCE | hRANKL2 | sOdf | tumor necrosis factor (ligand) superfamily, member 11 | - | HPRD | 10635328 |
| TRIB3 | C20orf97 | NIPK | SINK | SKIP3 | TRB3 | tribbles homolog 3 (Drosophila) | Affinity Capture-Western Reconstituted Complex | BioGRID | 12791994 |
| TSC1 | KIAA0243 | LAM | MGC86987 | TSC | tuberous sclerosis 1 | - | HPRD,BioGRID | 12167664 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | - | HPRD,BioGRID | 12167664 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | Akt interacts with and phosphorylates tuberin. This interaction was modelled on a demonstrated interaction between Akt from an unspecified species and human Tuberin. | BIND | 12150915 |
| UXS1 | FLJ23591 | UGD | UDP-glucuronate decarboxylase 1 | - | HPRD | 11877387 |
| YBX1 | BP-8 | CSDA2 | CSDB | DBPB | MDR-NF1 | MGC104858 | MGC110976 | MGC117250 | NSEP-1 | NSEP1 | YB-1 | YB1 | Y box binding protein 1 | Akt-1 interacts with and phosphorylates YB-1. | BIND | 15806160 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD,BioGRID | 11956222 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Akt interaction with and phosphorylation of 14-3-3 zeta at the same residue as MAPKAPK2. | BIND | 11956222 |